Yuli Wang
A multitask framework for automated interpretation of multi-frame right upper quadrant ultrasound in clinical decision support
Jan 17, 2026Abstract:Ultrasound is a cornerstone of emergency and hepatobiliary imaging, yet its interpretation remains highly operator-dependent and time-sensitive. Here, we present a multitask vision-language agent (VLM) developed to assist with comprehensive right upper quadrant (RUQ) ultrasound interpretation across the full diagnostic workflow. The system was trained on a large, multi-center dataset comprising a primary cohort from Johns Hopkins Medical Institutions (9,189 cases, 594,099 images) and externally validated on cohorts from Stanford University (108 cases, 3,240 images) and a major Chinese medical center (257 cases, 3,178 images). Built on the Qwen2.5-VL-7B architecture, the agent integrates frame-level visual understanding with report-grounded language reasoning to perform three tasks: (i) classification of 18 hepatobiliary and gallbladder conditions, (ii) generation of clinically coherent diagnostic reports, and (iii) surgical decision support based on ultrasound findings and clinical data. The model achieved high diagnostic accuracy across all tasks, generated reports that were indistinguishable from expert-written versions in blinded evaluations, and demonstrated superior factual accuracy and information density on content-based metrics. The agent further identified patients requiring cholecystectomy with high precision, supporting real-time decision-making. These results highlight the potential of generalist vision-language models to improve diagnostic consistency, reporting efficiency, and surgical triage in real-world ultrasound practice.
Dataset and Benchmark for Enhancing Critical Retained Foreign Object Detection
Jul 09, 2025Abstract:Critical retained foreign objects (RFOs), including surgical instruments like sponges and needles, pose serious patient safety risks and carry significant financial and legal implications for healthcare institutions. Detecting critical RFOs using artificial intelligence remains challenging due to their rarity and the limited availability of chest X-ray datasets that specifically feature critical RFOs cases. Existing datasets only contain non-critical RFOs, like necklace or zipper, further limiting their utility for developing clinically impactful detection algorithms. To address these limitations, we introduce "Hopkins RFOs Bench", the first and largest dataset of its kind, containing 144 chest X-ray images of critical RFO cases collected over 18 years from the Johns Hopkins Health System. Using this dataset, we benchmark several state-of-the-art object detection models, highlighting the need for enhanced detection methodologies for critical RFO cases. Recognizing data scarcity challenges, we further explore image synthetic methods to bridge this gap. We evaluate two advanced synthetic image methods, DeepDRR-RFO, a physics-based method, and RoentGen-RFO, a diffusion-based method, for creating realistic radiographs featuring critical RFOs. Our comprehensive analysis identifies the strengths and limitations of each synthetic method, providing insights into effectively utilizing synthetic data to enhance model training. The Hopkins RFOs Bench and our findings significantly advance the development of reliable, generalizable AI-driven solutions for detecting critical RFOs in clinical chest X-rays.
Abn-BLIP: Abnormality-aligned Bootstrapping Language-Image Pre-training for Pulmonary Embolism Diagnosis and Report Generation from CTPA
Mar 03, 2025Abstract:Medical imaging plays a pivotal role in modern healthcare, with computed tomography pulmonary angiography (CTPA) being a critical tool for diagnosing pulmonary embolism and other thoracic conditions. However, the complexity of interpreting CTPA scans and generating accurate radiology reports remains a significant challenge. This paper introduces Abn-BLIP (Abnormality-aligned Bootstrapping Language-Image Pretraining), an advanced diagnosis model designed to align abnormal findings to generate the accuracy and comprehensiveness of radiology reports. By leveraging learnable queries and cross-modal attention mechanisms, our model demonstrates superior performance in detecting abnormalities, reducing missed findings, and generating structured reports compared to existing methods. Our experiments show that Abn-BLIP outperforms state-of-the-art medical vision-language models and 3D report generation methods in both accuracy and clinical relevance. These results highlight the potential of integrating multimodal learning strategies for improving radiology reporting. The source code is available at https://github.com/zzs95/abn-blip.
Evaluating unsupervised contrastive learning framework for MRI sequences classification
Jan 12, 2025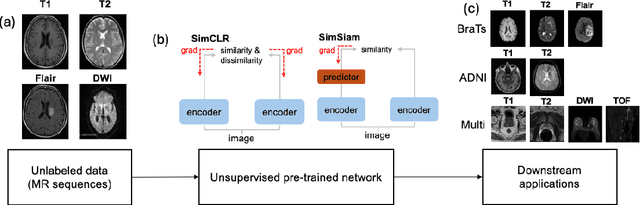
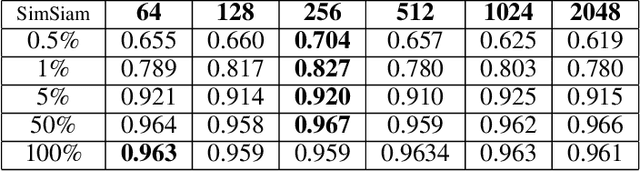
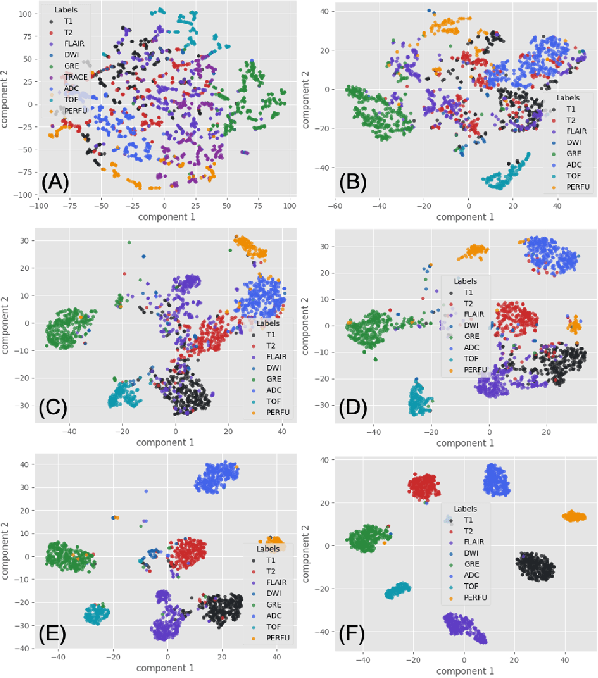
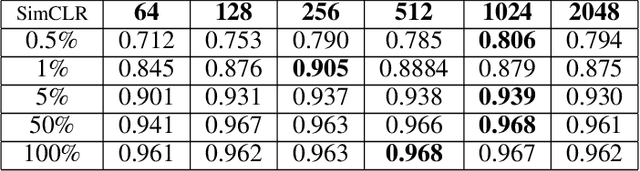
Abstract:The automatic identification of Magnetic Resonance Imaging (MRI) sequences can streamline clinical workflows by reducing the time radiologists spend manually sorting and identifying sequences, thereby enabling faster diagnosis and treatment planning for patients. However, the lack of standardization in the parameters of MRI scans poses challenges for automated systems and complicates the generation and utilization of datasets for machine learning research. To address this issue, we propose a system for MRI sequence identification using an unsupervised contrastive deep learning framework. By training a convolutional neural network based on the ResNet-18 architecture, our system classifies nine common MRI sequence types as a 9-class classification problem. The network was trained using an in-house internal dataset and validated on several public datasets, including BraTS, ADNI, Fused Radiology-Pathology Prostate Dataset, the Breast Cancer Dataset (ACRIN), among others, encompassing diverse acquisition protocols and requiring only 2D slices for training. Our system achieves a classification accuracy of over 0.95 across the nine most common MRI sequence types.
Optimizing Prompt Strategies for SAM: Advancing lesion Segmentation Across Diverse Medical Imaging Modalities
Dec 28, 2024



Abstract:Purpose: To evaluate various Segmental Anything Model (SAM) prompt strategies across four lesions datasets and to subsequently develop a reinforcement learning (RL) agent to optimize SAM prompt placement. Materials and Methods: This retrospective study included patients with four independent ovarian, lung, renal, and breast tumor datasets. Manual segmentation and SAM-assisted segmentation were performed for all lesions. A RL model was developed to predict and select SAM points to maximize segmentation performance. Statistical analysis of segmentation was conducted using pairwise t-tests. Results: Results show that increasing the number of prompt points significantly improves segmentation accuracy, with Dice coefficients rising from 0.272 for a single point to 0.806 for five or more points in ovarian tumors. The prompt location also influenced performance, with surface and union-based prompts outperforming center-based prompts, achieving mean Dice coefficients of 0.604 and 0.724 for ovarian and breast tumors, respectively. The RL agent achieved a peak Dice coefficient of 0.595 for ovarian tumors, outperforming random and alternative RL strategies. Additionally, it significantly reduced segmentation time, achieving a nearly 10-fold improvement compared to manual methods using SAM. Conclusion: While increased SAM prompts and non-centered prompts generally improved segmentation accuracy, each pathology and modality has specific optimal thresholds and placement strategies. Our RL agent achieved superior performance compared to other agents while achieving a significant reduction in segmentation time.
Optimizing prompt strategies for the Segment Anything Model are explored, focusing on prompt location, number, and reinforcement learning-based agent for prompt placement across four lesion datasets
Dec 23, 2024



Abstract:Purpose: To evaluate various Segmental Anything Model (SAM) prompt strategies across four lesions datasets and to subsequently develop a reinforcement learning (RL) agent to optimize SAM prompt placement. Materials and Methods: This retrospective study included patients with four independent ovarian, lung, renal, and breast tumor datasets. Manual segmentation and SAM-assisted segmentation were performed for all lesions. A RL model was developed to predict and select SAM points to maximize segmentation performance. Statistical analysis of segmentation was conducted using pairwise t-tests. Results: Results show that increasing the number of prompt points significantly improves segmentation accuracy, with Dice coefficients rising from 0.272 for a single point to 0.806 for five or more points in ovarian tumors. The prompt location also influenced performance, with surface and union-based prompts outperforming center-based prompts, achieving mean Dice coefficients of 0.604 and 0.724 for ovarian and breast tumors, respectively. The RL agent achieved a peak Dice coefficient of 0.595 for ovarian tumors, outperforming random and alternative RL strategies. Additionally, it significantly reduced segmentation time, achieving a nearly 10-fold improvement compared to manual methods using SAM. Conclusion: While increased SAM prompts and non-centered prompts generally improved segmentation accuracy, each pathology and modality has specific optimal thresholds and placement strategies. Our RL agent achieved superior performance compared to other agents while achieving a significant reduction in segmentation time.
Is Registering Raw Tagged-MR Enough for Strain Estimation in the Era of Deep Learning?
Jan 31, 2024Abstract:Magnetic Resonance Imaging with tagging (tMRI) has long been utilized for quantifying tissue motion and strain during deformation. However, a phenomenon known as tag fading, a gradual decrease in tag visibility over time, often complicates post-processing. The first contribution of this study is to model tag fading by considering the interplay between $T_1$ relaxation and the repeated application of radio frequency (RF) pulses during serial imaging sequences. This is a factor that has been overlooked in prior research on tMRI post-processing. Further, we have observed an emerging trend of utilizing raw tagged MRI within a deep learning-based (DL) registration framework for motion estimation. In this work, we evaluate and analyze the impact of commonly used image similarity objectives in training DL registrations on raw tMRI. This is then compared with the Harmonic Phase-based approach, a traditional approach which is claimed to be robust to tag fading. Our findings, derived from both simulated images and an actual phantom scan, reveal the limitations of various similarity losses in raw tMRI and emphasize caution in registration tasks where image intensity changes over time.
Efficient Annotation for Medical Image Analysis: A One-Pass Selective Annotation Approach
Sep 15, 2023



Abstract:Annotating biomedical images for supervised learning is a complex and labor-intensive task due to data diversity and its intricate nature. In this paper, we propose an innovative method, the efficient one-pass selective annotation (EPOSA), that significantly reduces the annotation burden while maintaining robust model performance. Our approach employs a variational autoencoder (VAE) to extract salient features from unannotated images, which are subsequently clustered using the DBSCAN algorithm. This process groups similar images together, forming distinct clusters. We then use a two-stage sample selection algorithm, called representative selection (RepSel), to form a selected dataset. The first stage is a Markov Chain Monte Carlo (MCMC) sampling technique to select representative samples from each cluster for annotations. This selection process is the second stage, which is guided by the principle of maximizing intra-cluster mutual information and minimizing inter-cluster mutual information. This ensures a diverse set of features for model training and minimizes outlier inclusion. The selected samples are used to train a VGG-16 network for image classification. Experimental results on the Med-MNIST dataset demonstrate that our proposed EPOSA outperforms random selection and other state-of-the-art methods under the same annotation budget, presenting a promising direction for efficient and effective annotation in medical image analysis.
Optimal operating MR contrast for brain ventricle parcellation
Apr 04, 2023
Abstract:Development of MR harmonization has enabled different contrast MRIs to be synthesized while preserving the underlying anatomy. In this paper, we use image harmonization to explore the impact of different T1-w MR contrasts on a state-of-the-art ventricle parcellation algorithm VParNet. We identify an optimal operating contrast (OOC) for ventricle parcellation; by showing that the performance of a pretrained VParNet can be boosted by adjusting contrast to the OOC.
Label Propagation via Random Walk for Training Robust Thalamus Nuclei Parcellation Model from Noisy Annotations
Mar 30, 2023Abstract:Data-driven thalamic nuclei parcellation depends on high-quality manual annotations. However, the small size and low contrast changes among thalamic nuclei, yield annotations that are often incomplete, noisy, or ambiguously labelled. To train a robust thalamic nuclei parcellation model with noisy annotations, we propose a label propagation algorithm based on random walker to refine the annotations before model training. A two-step model was trained to generate first the whole thalamus and then the nuclei masks. We conducted experiments on a mild traumatic brain injury~(mTBI) dataset with noisy thalamic nuclei annotations. Our model outperforms current state-of-the-art thalamic nuclei parcellations by a clear margin. We believe our method can also facilitate the training of other parcellation models with noisy labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge