Yuan Xue
LHAW: Controllable Underspecification for Long-Horizon Tasks
Feb 11, 2026Abstract:Long-horizon workflow agents that operate effectively over extended periods are essential for truly autonomous systems. Their reliable execution critically depends on the ability to reason through ambiguous situations in which clarification seeking is necessary to ensure correct task execution. However, progress is limited by the lack of scalable, task-agnostic frameworks for systematically curating and measuring the impact of ambiguity across custom workflows. We address this gap by introducing LHAW (Long-Horizon Augmented Workflows), a modular, dataset-agnostic synthetic pipeline that transforms any well-specified task into controllable underspecified variants by systematically removing information across four dimensions - Goals, Constraints, Inputs, and Context - at configurable severity levels. Unlike approaches that rely on LLM predictions of ambiguity, LHAW validates variants through empirical agent trials, classifying them as outcome-critical, divergent, or benign based on observed terminal state divergence. We release 285 task variants from TheAgentCompany, SWE-Bench Pro and MCP-Atlas according to our taxonomy alongside formal analysis measuring how current agents detect, reason about, and resolve underspecification across ambiguous settings. LHAW provides the first systematic framework for cost-sensitive evaluation of agent clarification behavior in long-horizon settings, enabling development of reliable autonomous systems.
Atlas is Your Perfect Context: One-Shot Customization for Generalizable Foundational Medical Image Segmentation
Dec 20, 2025



Abstract:Accurate medical image segmentation is essential for clinical diagnosis and treatment planning. While recent interactive foundation models (e.g., nnInteractive) enhance generalization through large-scale multimodal pretraining, they still depend on precise prompts and often perform below expectations in contexts that are underrepresented in their training data. We present AtlasSegFM, an atlas-guided framework that customizes available foundation models to clinical contexts with a single annotated example. The core innovations are: 1) a pipeline that provides context-aware prompts for foundation models via registration between a context atlas and query images, and 2) a test-time adapter to fuse predictions from both atlas registration and the foundation model. Extensive experiments across public and in-house datasets spanning multiple modalities and organs demonstrate that AtlasSegFM consistently improves segmentation, particularly for small, delicate structures. AtlasSegFM provides a lightweight, deployable solution one-shot customization of foundation models in real-world clinical workflows. The code will be made publicly available.
LimiX: Unleashing Structured-Data Modeling Capability for Generalist Intelligence
Sep 03, 2025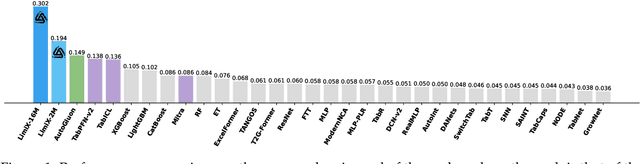
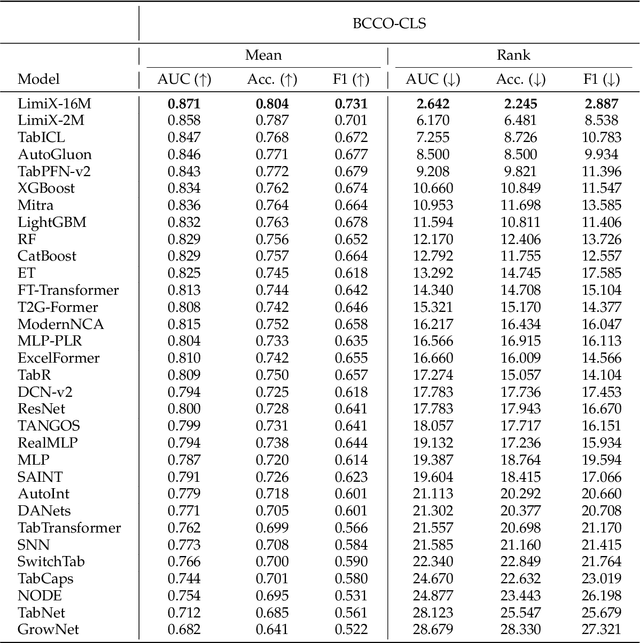
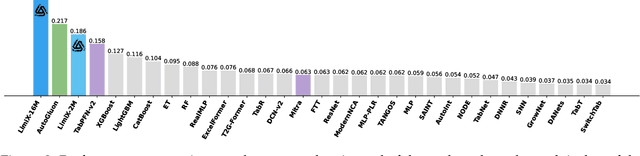
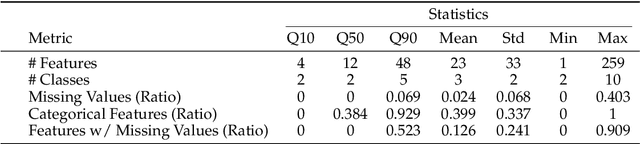
Abstract:We argue that progress toward general intelligence requires complementary foundation models grounded in language, the physical world, and structured data. This report presents LimiX, the first installment of our large structured-data models (LDMs). LimiX treats structured data as a joint distribution over variables and missingness, thus capable of addressing a wide range of tabular tasks through query-based conditional prediction via a single model. LimiX is pretrained using masked joint-distribution modeling with an episodic, context-conditional objective, where the model predicts for query subsets conditioned on dataset-specific contexts, supporting rapid, training-free adaptation at inference. We evaluate LimiX across 10 large structured-data benchmarks with broad regimes of sample size, feature dimensionality, class number, categorical-to-numerical feature ratio, missingness, and sample-to-feature ratios. With a single model and a unified interface, LimiX consistently surpasses strong baselines including gradient-boosting trees, deep tabular networks, recent tabular foundation models, and automated ensembles, as shown in Figure 1 and Figure 2. The superiority holds across a wide range of tasks, such as classification, regression, missing value imputation, and data generation, often by substantial margins, while avoiding task-specific architectures or bespoke training per task. All LimiX models are publicly accessible under Apache 2.0.
Feature-Based Instance Neighbor Discovery: Advanced Stable Test-Time Adaptation in Dynamic World
Jun 07, 2025Abstract:Despite progress, deep neural networks still suffer performance declines under distribution shifts between training and test domains, leading to a substantial decrease in Quality of Experience (QoE) for applications. Existing test-time adaptation (TTA) methods are challenged by dynamic, multiple test distributions within batches. We observe that feature distributions across different domains inherently cluster into distinct groups with varying means and variances. This divergence reveals a critical limitation of previous global normalization strategies in TTA, which inevitably distort the original data characteristics. Based on this insight, we propose Feature-based Instance Neighbor Discovery (FIND), which comprises three key components: Layer-wise Feature Disentanglement (LFD), Feature Aware Batch Normalization (FABN) and Selective FABN (S-FABN). LFD stably captures features with similar distributions at each layer by constructing graph structures. While FABN optimally combines source statistics with test-time distribution specific statistics for robust feature representation. Finally, S-FABN determines which layers require feature partitioning and which can remain unified, thereby enhancing inference efficiency. Extensive experiments demonstrate that FIND significantly outperforms existing methods, achieving a 30\% accuracy improvement in dynamic scenarios while maintaining computational efficiency.
Computer-Aided Layout Generation for Building Design: A Review
Apr 13, 2025
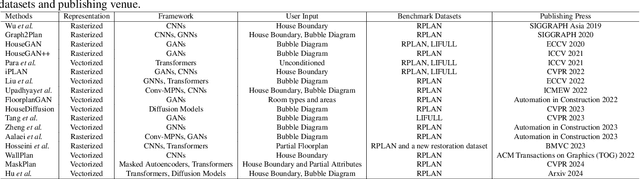
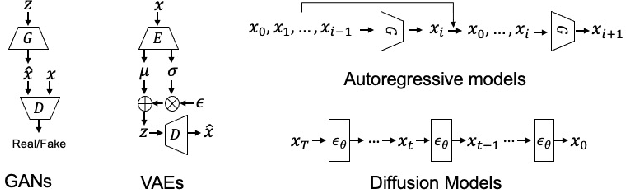

Abstract:Generating realistic building layouts for automatic building design has been studied in both the computer vision and architecture domains. Traditional approaches from the architecture domain, which are based on optimization techniques or heuristic design guidelines, can synthesize desirable layouts, but usually require post-processing and involve human interaction in the design pipeline, making them costly and timeconsuming. The advent of deep generative models has significantly improved the fidelity and diversity of the generated architecture layouts, reducing the workload by designers and making the process much more efficient. In this paper, we conduct a comprehensive review of three major research topics of architecture layout design and generation: floorplan layout generation, scene layout synthesis, and generation of some other formats of building layouts. For each topic, we present an overview of the leading paradigms, categorized either by research domains (architecture or machine learning) or by user input conditions or constraints. We then introduce the commonly-adopted benchmark datasets that are used to verify the effectiveness of the methods, as well as the corresponding evaluation metrics. Finally, we identify the well-solved problems and limitations of existing approaches, then propose new perspectives as promising directions for future research in this important research area. A project associated with this survey to maintain the resources is available at awesome-building-layout-generation.
MATHGLANCE: Multimodal Large Language Models Do Not Know Where to Look in Mathematical Diagrams
Mar 26, 2025Abstract:Diagrams serve as a fundamental form of visual language, representing complex concepts and their inter-relationships through structured symbols, shapes, and spatial arrangements. Unlike natural images, their inherently symbolic and abstract nature poses significant challenges for Multimodal Large Language Models (MLLMs). However, current benchmarks conflate perceptual and reasoning tasks, making it difficult to assess whether MLLMs genuinely understand mathematical diagrams beyond superficial pattern recognition. To address this gap, we introduce MATHGLANCE, a benchmark specifically designed to isolate and evaluate mathematical perception in MLLMs. MATHGLANCE comprises 1.2K images and 1.6K carefully curated questions spanning four perception tasks: shape classification, object counting, relationship identification, and object grounding, covering diverse domains including plane geometry, solid geometry, and graphical representations. Our evaluation of MLLMs reveals that their ability to understand diagrams is notably limited, particularly in fine-grained grounding tasks. In response, we construct GeoPeP, a perception-oriented dataset of 200K structured geometry image-text pairs explicitly annotated with geometric primitives and precise spatial relationships. Training MLLM on GeoPeP leads to significant gains in perceptual accuracy, which in turn substantially improves mathematical reasoning. Our benchmark and dataset establish critical standards for evaluating and advancing multimodal mathematical understanding, providing valuable resources and insights to foster future MLLM research.
Understanding the Generalization of In-Context Learning in Transformers: An Empirical Study
Mar 19, 2025Abstract:Large language models (LLMs) like GPT-4 and LLaMA-3 utilize the powerful in-context learning (ICL) capability of Transformer architecture to learn on the fly from limited examples. While ICL underpins many LLM applications, its full potential remains hindered by a limited understanding of its generalization boundaries and vulnerabilities. We present a systematic investigation of transformers' generalization capability with ICL relative to training data coverage by defining a task-centric framework along three dimensions: inter-problem, intra-problem, and intra-task generalization. Through extensive simulation and real-world experiments, encompassing tasks such as function fitting, API calling, and translation, we find that transformers lack inter-problem generalization with ICL, but excel in intra-task and intra-problem generalization. When the training data includes a greater variety of mixed tasks, it significantly enhances the generalization ability of ICL on unseen tasks and even on known simple tasks. This guides us in designing training data to maximize the diversity of tasks covered and to combine different tasks whenever possible, rather than solely focusing on the target task for testing.
Open Eyes, Then Reason: Fine-grained Visual Mathematical Understanding in MLLMs
Jan 11, 2025



Abstract:Current multimodal large language models (MLLMs) often underperform on mathematical problem-solving tasks that require fine-grained visual understanding. The limitation is largely attributable to inadequate perception of geometric primitives during image-level contrastive pre-training (e.g., CLIP). While recent efforts to improve math MLLMs have focused on scaling up mathematical visual instruction datasets and employing stronger LLM backbones, they often overlook persistent errors in visual recognition. In this paper, we systematically evaluate the visual grounding capabilities of state-of-the-art MLLMs and reveal a significant negative correlation between visual grounding accuracy and problem-solving performance, underscoring the critical role of fine-grained visual understanding. Notably, advanced models like GPT-4o exhibit a 70% error rate when identifying geometric entities, highlighting that this remains a key bottleneck in visual mathematical reasoning. To address this, we propose a novel approach, SVE-Math (Selective Vision-Enhanced Mathematical MLLM), featuring a geometric-grounded vision encoder and a feature router that dynamically adjusts the contribution of hierarchical visual feature maps. Our model recognizes accurate visual primitives and generates precise visual prompts tailored to the language model's reasoning needs. In experiments, SVE-Math-Qwen2.5-7B outperforms other 7B models by 15% on MathVerse and is compatible with GPT-4V on MathVista. Despite being trained on smaller datasets, SVE-Math-7B achieves competitive performance on GeoQA, rivaling models trained on significantly larger datasets. Our findings emphasize the importance of incorporating fine-grained visual understanding into MLLMs and provide a promising direction for future research.
LL-ICM: Image Compression for Low-level Machine Vision via Large Vision-Language Model
Dec 05, 2024



Abstract:Image Compression for Machines (ICM) aims to compress images for machine vision tasks rather than human viewing. Current works predominantly concentrate on high-level tasks like object detection and semantic segmentation. However, the quality of original images is usually not guaranteed in the real world, leading to even worse perceptual quality or downstream task performance after compression. Low-level (LL) machine vision models, like image restoration models, can help improve such quality, and thereby their compression requirements should also be considered. In this paper, we propose a pioneered ICM framework for LL machine vision tasks, namely LL-ICM. By jointly optimizing compression and LL tasks, the proposed LL-ICM not only enriches its encoding ability in generalizing to versatile LL tasks but also optimizes the processing ability of down-stream LL task models, achieving mutual adaptation for image codecs and LL task models. Furthermore, we integrate large-scale vision-language models into the LL-ICM framework to generate more universal and distortion-robust feature embeddings for LL vision tasks. Therefore, one LL-ICM codec can generalize to multiple tasks. We establish a solid benchmark to evaluate LL-ICM, which includes extensive objective experiments by using both full and no-reference image quality assessments. Experimental results show that LL-ICM can achieve 22.65% BD-rate reductions over the state-of-the-art methods.
A MEMS-based terahertz broadband beam steering technique
Sep 06, 2024Abstract:A multi-level tunable reflection array wide-angle beam scanning method is proposed to address the limited bandwidth and small scanning angle issues of current terahertz beam scanning technology. In this method, a focusing lens and its array are used to achieve terahertz wave spatial beam control, and MEMS mirrors and their arrays are used to achieve wide-angle beam scanning. The 1~3 order terahertz MEMS beam scanning system designed based on this method can extend the mechanical scanning angle of MEMS mirrors by 2~6 times, when tested and verified using an electromagnetic MEMS mirror with a 7mm optical aperture and a scanning angle of 15{\deg} and a D-band terahertz signal source. The experiment shows that the operating bandwidth of the first-order terahertz MEMS beam scanning system is better than 40GHz, the continuous beam scanning angle is about 30{\deg}, the continuous beam scanning cycle response time is about 1.1ms, and the antenna gain is better than 15dBi at 160GHz. This method has been validated for its large bandwidth and scalable scanning angle, and has potential application prospects in terahertz dynamic communication, detection radar, scanning imaging, and other fields.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge