Chuyang Ye
Beijing Institute of Technology
MolCLIP: A Molecular-Auxiliary CLIP Framework for Identifying Drug Mechanism of Action Based on Time-Lapsed Mitochondrial Images
Jul 10, 2025



Abstract:Drug Mechanism of Action (MoA) mainly investigates how drug molecules interact with cells, which is crucial for drug discovery and clinical application. Recently, deep learning models have been used to recognize MoA by relying on high-content and fluorescence images of cells exposed to various drugs. However, these methods focus on spatial characteristics while overlooking the temporal dynamics of live cells. Time-lapse imaging is more suitable for observing the cell response to drugs. Additionally, drug molecules can trigger cellular dynamic variations related to specific MoA. This indicates that the drug molecule modality may complement the image counterpart. This paper proposes MolCLIP, the first visual language model to combine microscopic cell video- and molecule-modalities. MolCLIP designs a molecule-auxiliary CLIP framework to guide video features in learning the distribution of the molecular latent space. Furthermore, we integrate a metric learning strategy with MolCLIP to optimize the aggregation of video features. Experimental results on the MitoDataset demonstrate that MolCLIP achieves improvements of 51.2% and 20.5% in mAP for drug identification and MoA recognition, respectively.
Feature-Based Instance Neighbor Discovery: Advanced Stable Test-Time Adaptation in Dynamic World
Jun 07, 2025Abstract:Despite progress, deep neural networks still suffer performance declines under distribution shifts between training and test domains, leading to a substantial decrease in Quality of Experience (QoE) for applications. Existing test-time adaptation (TTA) methods are challenged by dynamic, multiple test distributions within batches. We observe that feature distributions across different domains inherently cluster into distinct groups with varying means and variances. This divergence reveals a critical limitation of previous global normalization strategies in TTA, which inevitably distort the original data characteristics. Based on this insight, we propose Feature-based Instance Neighbor Discovery (FIND), which comprises three key components: Layer-wise Feature Disentanglement (LFD), Feature Aware Batch Normalization (FABN) and Selective FABN (S-FABN). LFD stably captures features with similar distributions at each layer by constructing graph structures. While FABN optimally combines source statistics with test-time distribution specific statistics for robust feature representation. Finally, S-FABN determines which layers require feature partitioning and which can remain unified, thereby enhancing inference efficiency. Extensive experiments demonstrate that FIND significantly outperforms existing methods, achieving a 30\% accuracy improvement in dynamic scenarios while maintaining computational efficiency.
DATTA: Towards Diversity Adaptive Test-Time Adaptation in Dynamic Wild World
Aug 15, 2024
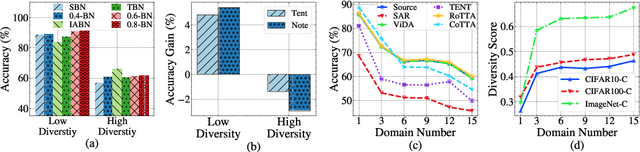
Abstract:Test-time adaptation (TTA) effectively addresses distribution shifts between training and testing data by adjusting models on test samples, which is crucial for improving model inference in real-world applications. However, traditional TTA methods typically follow a fixed pattern to address the dynamic data patterns (low-diversity or high-diversity patterns) often leading to performance degradation and consequently a decline in Quality of Experience (QoE). The primary issues we observed are:Different scenarios require different normalization methods (e.g., Instance Normalization is optimal in mixed domains but not in static domains). Model fine-tuning can potentially harm the model and waste time.Hence, it is crucial to design strategies for effectively measuring and managing distribution diversity to minimize its negative impact on model performance. Based on these observations, this paper proposes a new general method, named Diversity Adaptive Test-Time Adaptation (DATTA), aimed at improving QoE. DATTA dynamically selects the best batch normalization methods and fine-tuning strategies by leveraging the Diversity Score to differentiate between high and low diversity score batches. It features three key components: Diversity Discrimination (DD) to assess batch diversity, Diversity Adaptive Batch Normalization (DABN) to tailor normalization methods based on DD insights, and Diversity Adaptive Fine-Tuning (DAFT) to selectively fine-tune the model. Experimental results show that our method achieves up to a 21% increase in accuracy compared to state-of-the-art methodologies, indicating that our method maintains good model performance while demonstrating its robustness. Our code will be released soon.
CAVM: Conditional Autoregressive Vision Model for Contrast-Enhanced Brain Tumor MRI Synthesis
Jun 23, 2024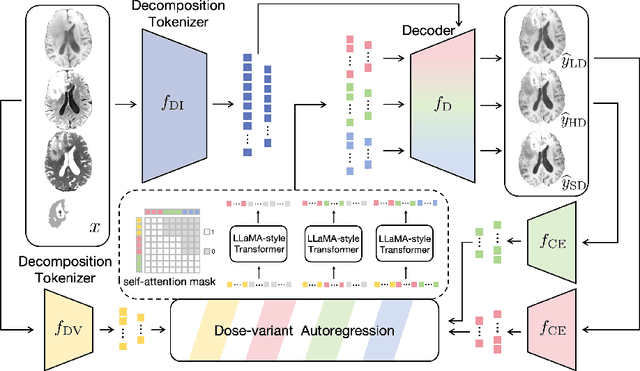
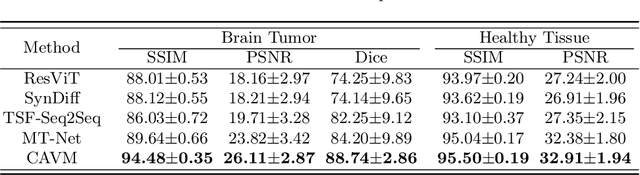
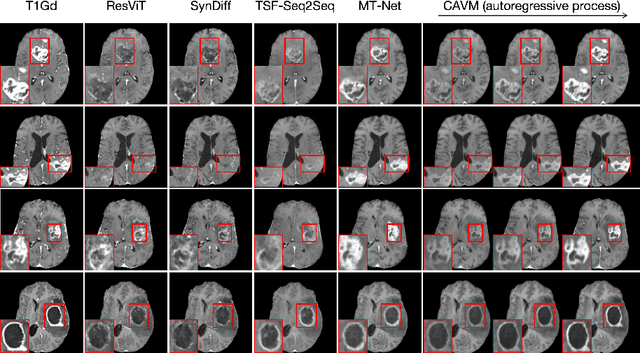

Abstract:Contrast-enhanced magnetic resonance imaging (MRI) is pivotal in the pipeline of brain tumor segmentation and analysis. Gadolinium-based contrast agents, as the most commonly used contrast agents, are expensive and may have potential side effects, and it is desired to obtain contrast-enhanced brain tumor MRI scans without the actual use of contrast agents. Deep learning methods have been applied to synthesize virtual contrast-enhanced MRI scans from non-contrast images. However, as this synthesis problem is inherently ill-posed, these methods fall short in producing high-quality results. In this work, we propose Conditional Autoregressive Vision Model (CAVM) for improving the synthesis of contrast-enhanced brain tumor MRI. As the enhancement of image intensity grows with a higher dose of contrast agents, we assume that it is less challenging to synthesize a virtual image with a lower dose, where the difference between the contrast-enhanced and non-contrast images is smaller. Thus, CAVM gradually increases the contrast agent dosage and produces higher-dose images based on previous lower-dose ones until the final desired dose is achieved. Inspired by the resemblance between the gradual dose increase and the Chain-of-Thought approach in natural language processing, CAVM uses an autoregressive strategy with a decomposition tokenizer and a decoder. Specifically, the tokenizer is applied to obtain a more compact image representation for computational efficiency, and it decomposes the image into dose-variant and dose-invariant tokens. Then, a masked self-attention mechanism is developed for autoregression that gradually increases the dose of the virtual image based on the dose-variant tokens. Finally, the updated dose-variant tokens corresponding to the desired dose are decoded together with dose-invariant tokens to produce the final contrast-enhanced MRI.
Discover Your Neighbors: Advanced Stable Test-Time Adaptation in Dynamic World
Jun 08, 2024Abstract:Despite progress, deep neural networks still suffer performance declines under distribution shifts between training and test domains, leading to a substantial decrease in Quality of Experience (QoE) for multimedia applications. Existing test-time adaptation (TTA) methods are challenged by dynamic, multiple test distributions within batches. This work provides a new perspective on analyzing batch normalization techniques through class-related and class-irrelevant features, our observations reveal combining source and test batch normalization statistics robustly characterizes target distributions. However, test statistics must have high similarity. We thus propose Discover Your Neighbours (DYN), the first backward-free approach specialized for dynamic TTA. The core innovation is identifying similar samples via instance normalization statistics and clustering into groups which provides consistent class-irrelevant representations. Specifically, Our DYN consists of layer-wise instance statistics clustering (LISC) and cluster-aware batch normalization (CABN). In LISC, we perform layer-wise clustering of approximate feature samples at each BN layer by calculating the cosine similarity of instance normalization statistics across the batch. CABN then aggregates SBN and TCN statistics to collaboratively characterize the target distribution, enabling more robust representations. Experimental results validate DYN's robustness and effectiveness, demonstrating maintained performance under dynamic data stream patterns.
A Foundation Model for Brain Lesion Segmentation with Mixture of Modality Experts
May 16, 2024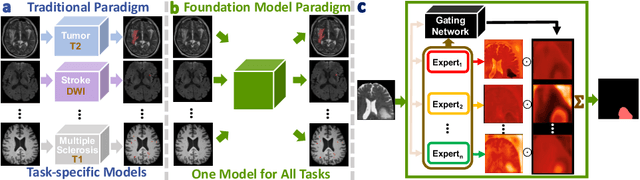
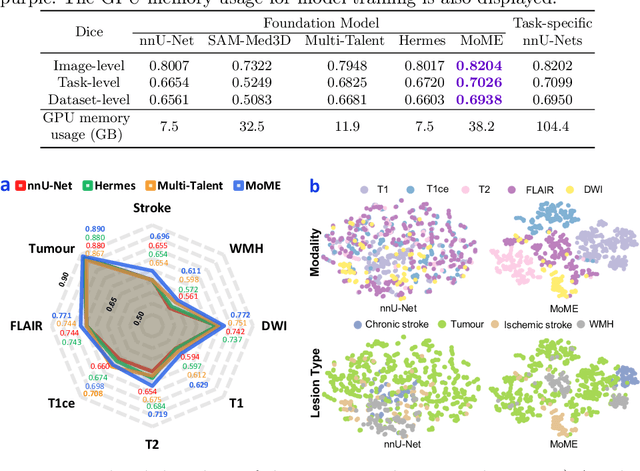

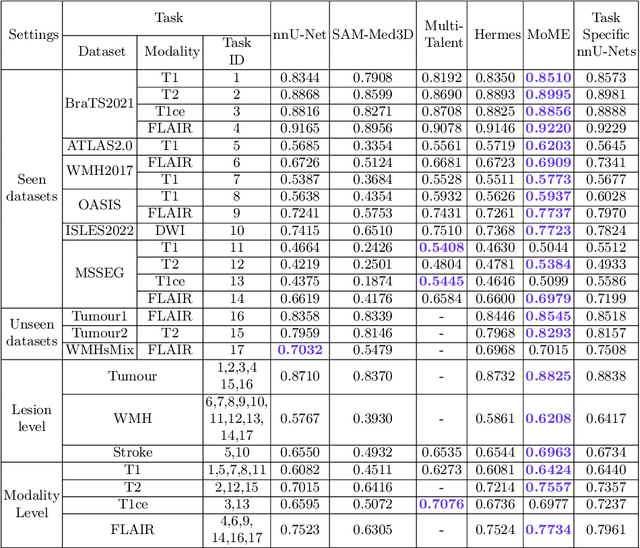
Abstract:Brain lesion segmentation plays an essential role in neurological research and diagnosis. As brain lesions can be caused by various pathological alterations, different types of brain lesions tend to manifest with different characteristics on different imaging modalities. Due to this complexity, brain lesion segmentation methods are often developed in a task-specific manner. A specific segmentation model is developed for a particular lesion type and imaging modality. However, the use of task-specific models requires predetermination of the lesion type and imaging modality, which complicates their deployment in real-world scenarios. In this work, we propose a universal foundation model for 3D brain lesion segmentation, which can automatically segment different types of brain lesions for input data of various imaging modalities. We formulate a novel Mixture of Modality Experts (MoME) framework with multiple expert networks attending to different imaging modalities. A hierarchical gating network combines the expert predictions and fosters expertise collaboration. Furthermore, we introduce a curriculum learning strategy during training to avoid the degeneration of each expert network and preserve their specialization. We evaluated the proposed method on nine brain lesion datasets, encompassing five imaging modalities and eight lesion types. The results show that our model outperforms state-of-the-art universal models and provides promising generalization to unseen datasets.
Advancing Brain Tumor Inpainting with Generative Models
Feb 02, 2024Abstract:Synthesizing healthy brain scans from diseased brain scans offers a potential solution to address the limitations of general-purpose algorithms, such as tissue segmentation and brain extraction algorithms, which may not effectively handle diseased images. We consider this a 3D inpainting task and investigate the adaptation of 2D inpainting methods to meet the requirements of 3D magnetic resonance imaging(MRI) data. Our contributions encompass potential modifications tailored to MRI-specific needs, and we conducted evaluations of multiple inpainting techniques using the BraTS2023 Inpainting datasets to assess their efficacy and limitations.
Better Generalization of White Matter Tract Segmentation to Arbitrary Datasets with Scaled Residual Bootstrap
Sep 25, 2023Abstract:White matter (WM) tract segmentation is a crucial step for brain connectivity studies. It is performed on diffusion magnetic resonance imaging (dMRI), and deep neural networks (DNNs) have achieved promising segmentation accuracy. Existing DNN-based methods use an annotated dataset for model training. However, the performance of the trained model on a different test dataset may not be optimal due to distribution shift, and it is desirable to design WM tract segmentation approaches that allow better generalization of the segmentation model to arbitrary test datasets. In this work, we propose a WM tract segmentation approach that improves the generalization with scaled residual bootstrap. The difference between dMRI scans in training and test datasets is most noticeably caused by the different numbers of diffusion gradients and noise levels. Since both of them lead to different signal-to-noise ratios (SNRs) between the training and test data, we propose to augment the training scans by adjusting the noise magnitude and develop an adapted residual bootstrap strategy for the augmentation. To validate the proposed approach, two dMRI datasets were used, and the experimental results show that our method consistently improved the generalization of WM tract segmentation under various settings.
Unsupervised Brain Tumor Segmentation with Image-based Prompts
Apr 04, 2023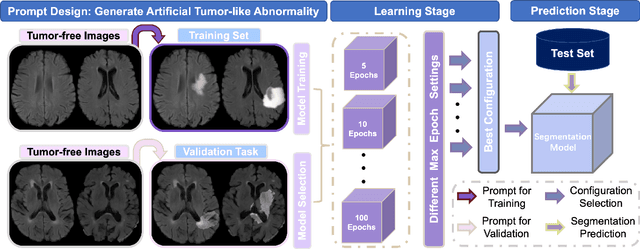
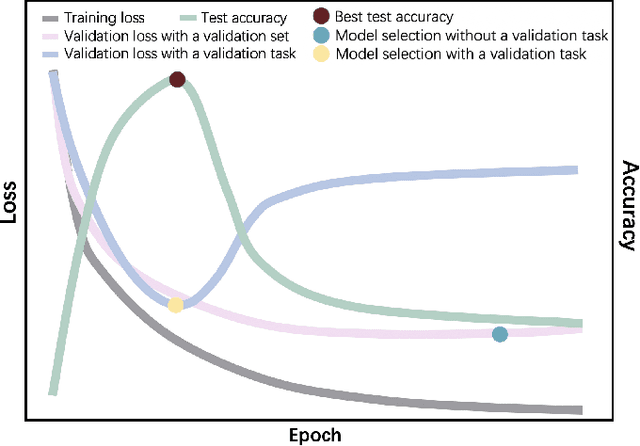
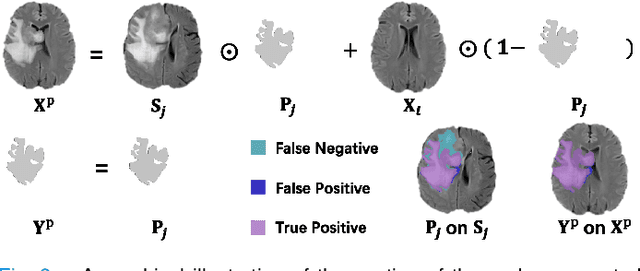
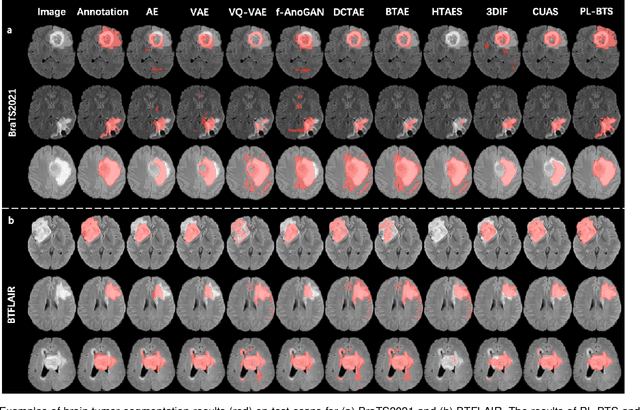
Abstract:Automated brain tumor segmentation based on deep learning (DL) has achieved promising performance. However, it generally relies on annotated images for model training, which is not always feasible in clinical settings. Therefore, the development of unsupervised DL-based brain tumor segmentation approaches without expert annotations is desired. Motivated by the success of prompt learning (PL) in natural language processing, we propose an approach to unsupervised brain tumor segmentation by designing image-based prompts that allow indication of brain tumors, and this approach is dubbed as PL-based Brain Tumor Segmentation (PL-BTS). Specifically, instead of directly training a model for brain tumor segmentation with a large amount of annotated data, we seek to train a model that can answer the question: is a voxel in the input image associated with tumor-like hyper-/hypo-intensity? Such a model can be trained by artificially generating tumor-like hyper-/hypo-intensity on images without tumors with hand-crafted designs. Since the hand-crafted designs may be too simplistic to represent all kinds of real tumors, the trained model may overfit the simplistic hand-crafted task rather than actually answer the question of abnormality. To address this problem, we propose the use of a validation task, where we generate a different hand-crafted task to monitor overfitting. In addition, we propose PL-BTS+ that further improves PL-BTS by exploiting unannotated images with brain tumors. Compared with competing unsupervised methods, the proposed method has achieved marked improvements on both public and in-house datasets, and we have also demonstrated its possible extension to other brain lesion segmentation tasks.
Fiber Tract Shape Measures Inform Prediction of Non-Imaging Phenotypes
Mar 16, 2023



Abstract:Neuroimaging measures of the brain's white matter connections can enable the prediction of non-imaging phenotypes, such as demographic and cognitive measures. Existing works have investigated traditional microstructure and connectivity measures from diffusion MRI tractography, without considering the shape of the connections reconstructed by tractography. In this paper, we investigate the potential of fiber tract shape features for predicting non-imaging phenotypes, both individually and in combination with traditional features. We focus on three basic shape features: length, diameter, and elongation. Two different prediction methods are used, including a traditional regression method and a deep-learning-based prediction method. Experiments use an efficient two-stage fusion strategy for prediction using microstructure, connectivity, and shape measures. To reduce predictive bias due to brain size, normalized shape features are also investigated. Experimental results on the Human Connectome Project (HCP) young adult dataset (n=1065) demonstrate that individual shape features are predictive of non-imaging phenotypes. When combined with microstructure and connectivity features, shape features significantly improve performance for predicting the cognitive score TPVT (NIH Toolbox picture vocabulary test). Overall, this study demonstrates that the shape of fiber tracts contains useful information for the description and study of the living human brain using machine learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge