Xinru Zhang
A Knowledge Noise Mitigation Framework for Knowledge-based Visual Question Answering
Sep 11, 2025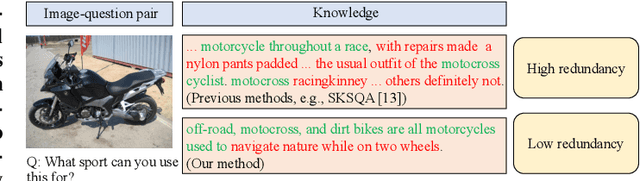
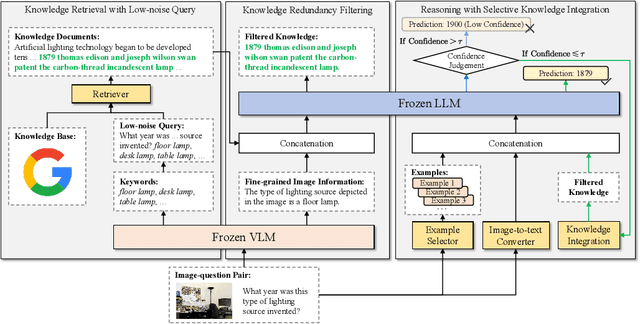
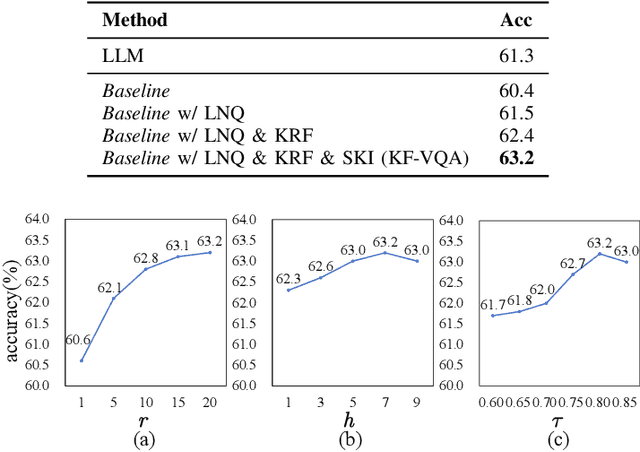
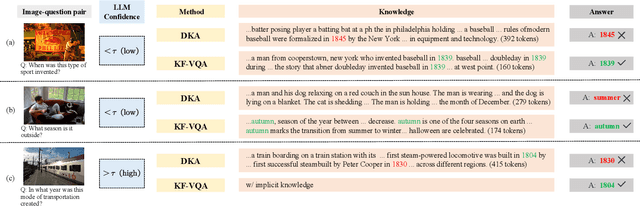
Abstract:Knowledge-based visual question answering (KB-VQA) requires a model to understand images and utilize external knowledge to provide accurate answers. Existing approaches often directly augment models with retrieved information from knowledge sources while ignoring substantial knowledge redundancy, which introduces noise into the answering process. To address this, we propose a training-free framework with knowledge focusing for KB-VQA, that mitigates the impact of noise by enhancing knowledge relevance and reducing redundancy. First, for knowledge retrieval, our framework concludes essential parts from the image-question pairs, creating low-noise queries that enhance the retrieval of highly relevant knowledge. Considering that redundancy still persists in the retrieved knowledge, we then prompt large models to identify and extract answer-beneficial segments from knowledge. In addition, we introduce a selective knowledge integration strategy, allowing the model to incorporate knowledge only when it lacks confidence in answering the question, thereby mitigating the influence of redundant information. Our framework enables the acquisition of accurate and critical knowledge, and extensive experiments demonstrate that it outperforms state-of-the-art methods.
Iterative Camera-LiDAR Extrinsic Optimization via Surrogate Diffusion
Nov 17, 2024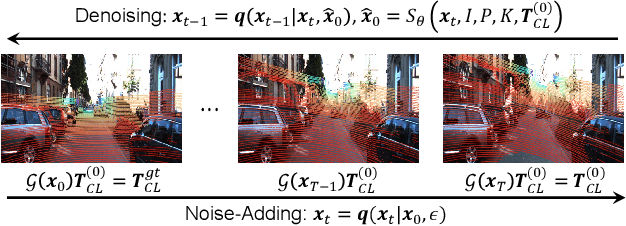
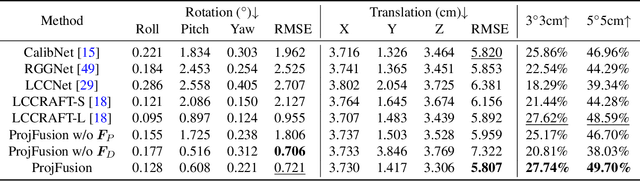
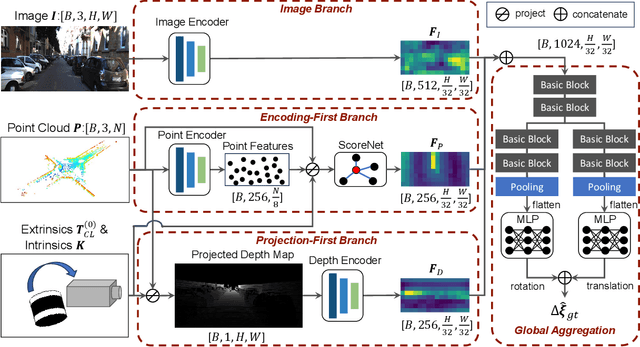
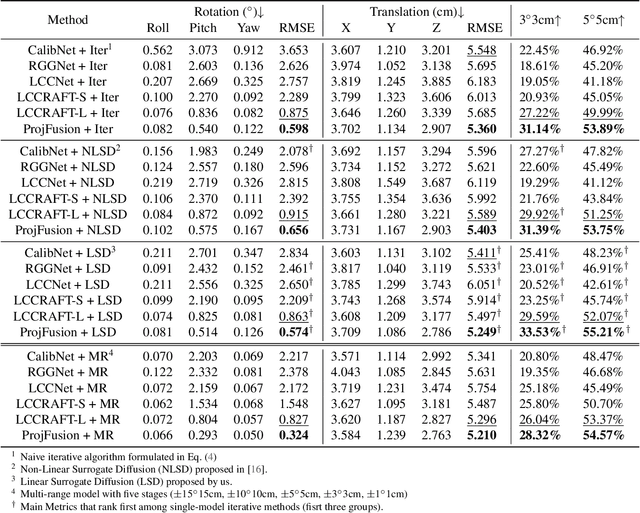
Abstract:Cameras and LiDAR are essential sensors for autonomous vehicles. Camera-LiDAR data fusion compensate for deficiencies of stand-alone sensors but relies on precise extrinsic calibration. Many learning-based calibration methods predict extrinsic parameters in a single step. Driven by the growing demand for higher accuracy, a few approaches utilize multi-range models or integrate multiple methods to improve extrinsic parameter predictions, but these strategies incur extended training times and require additional storage for separate models. To address these issues, we propose a single-model iterative approach based on surrogate diffusion to significantly enhance the capacity of individual calibration methods. By applying a buffering technique proposed by us, the inference time of our surrogate diffusion is 43.7% less than that of multi-range models. Additionally, we create a calibration network as our denoiser, featuring both projection-first and encoding-first branches for effective point feature extraction. Extensive experiments demonstrate that our diffusion model outperforms other single-model iterative methods and delivers competitive results compared to multi-range models. Our denoiser exceeds state-of-the-art calibration methods, reducing the rotation error by 24.5% compared to the second-best method. Furthermore, with the proposed diffusion applied, it achieves 20.4% less rotation error and 9.6% less translation error.
SegHeD: Segmentation of Heterogeneous Data for Multiple Sclerosis Lesions with Anatomical Constraints
Oct 02, 2024Abstract:Assessment of lesions and their longitudinal progression from brain magnetic resonance (MR) images plays a crucial role in diagnosing and monitoring multiple sclerosis (MS). Machine learning models have demonstrated a great potential for automated MS lesion segmentation. Training such models typically requires large-scale high-quality datasets that are consistently annotated. However, MS imaging datasets are often small, segregated across multiple sites, with different formats (cross-sectional or longitudinal), and diverse annotation styles. This poses a significant challenge to train a unified MS lesion segmentation model. To tackle this challenge, we present SegHeD, a novel multi-dataset multi-task segmentation model that can incorporate heterogeneous data as input and perform all-lesion, new-lesion, as well as vanishing-lesion segmentation. Furthermore, we account for domain knowledge about MS lesions, incorporating longitudinal, spatial, and volumetric constraints into the segmentation model. SegHeD is assessed on five MS datasets and achieves a high performance in all, new, and vanishing-lesion segmentation, outperforming several state-of-the-art methods in this field.
A Foundation Model for Brain Lesion Segmentation with Mixture of Modality Experts
May 16, 2024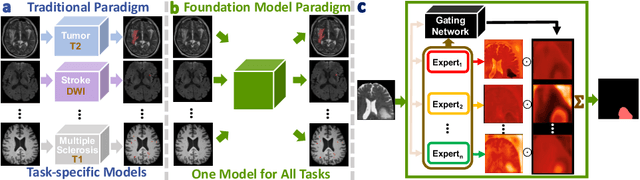
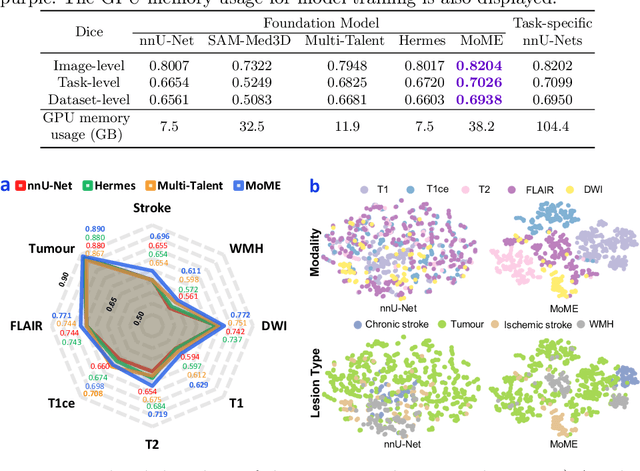

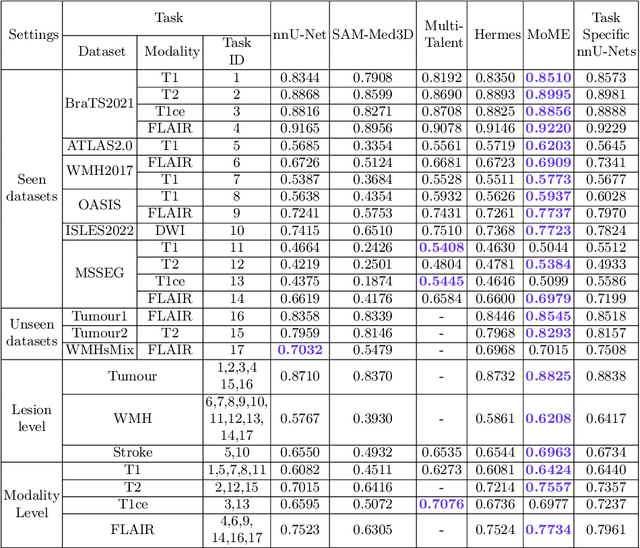
Abstract:Brain lesion segmentation plays an essential role in neurological research and diagnosis. As brain lesions can be caused by various pathological alterations, different types of brain lesions tend to manifest with different characteristics on different imaging modalities. Due to this complexity, brain lesion segmentation methods are often developed in a task-specific manner. A specific segmentation model is developed for a particular lesion type and imaging modality. However, the use of task-specific models requires predetermination of the lesion type and imaging modality, which complicates their deployment in real-world scenarios. In this work, we propose a universal foundation model for 3D brain lesion segmentation, which can automatically segment different types of brain lesions for input data of various imaging modalities. We formulate a novel Mixture of Modality Experts (MoME) framework with multiple expert networks attending to different imaging modalities. A hierarchical gating network combines the expert predictions and fosters expertise collaboration. Furthermore, we introduce a curriculum learning strategy during training to avoid the degeneration of each expert network and preserve their specialization. We evaluated the proposed method on nine brain lesion datasets, encompassing five imaging modalities and eight lesion types. The results show that our model outperforms state-of-the-art universal models and provides promising generalization to unseen datasets.
Advancing Brain Tumor Inpainting with Generative Models
Feb 02, 2024Abstract:Synthesizing healthy brain scans from diseased brain scans offers a potential solution to address the limitations of general-purpose algorithms, such as tissue segmentation and brain extraction algorithms, which may not effectively handle diseased images. We consider this a 3D inpainting task and investigate the adaptation of 2D inpainting methods to meet the requirements of 3D magnetic resonance imaging(MRI) data. Our contributions encompass potential modifications tailored to MRI-specific needs, and we conducted evaluations of multiple inpainting techniques using the BraTS2023 Inpainting datasets to assess their efficacy and limitations.
Unsupervised Brain Tumor Segmentation with Image-based Prompts
Apr 04, 2023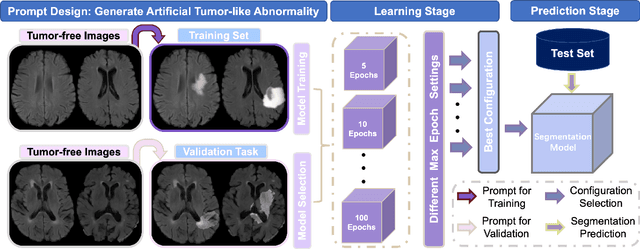
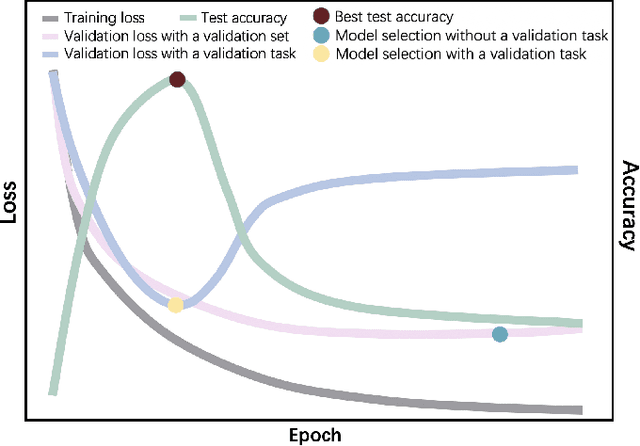
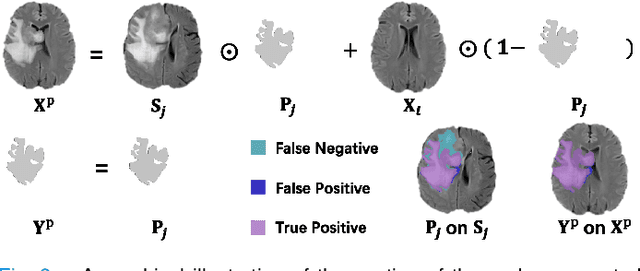
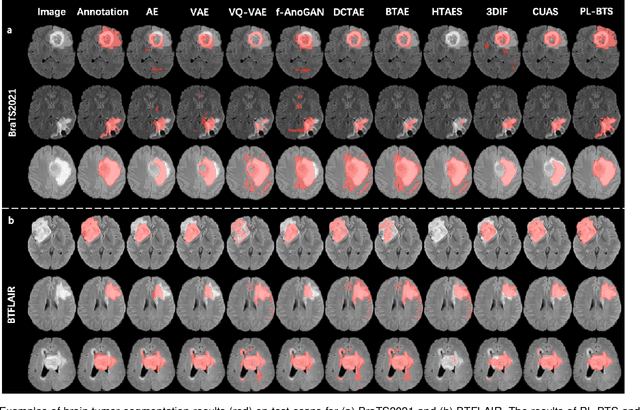
Abstract:Automated brain tumor segmentation based on deep learning (DL) has achieved promising performance. However, it generally relies on annotated images for model training, which is not always feasible in clinical settings. Therefore, the development of unsupervised DL-based brain tumor segmentation approaches without expert annotations is desired. Motivated by the success of prompt learning (PL) in natural language processing, we propose an approach to unsupervised brain tumor segmentation by designing image-based prompts that allow indication of brain tumors, and this approach is dubbed as PL-based Brain Tumor Segmentation (PL-BTS). Specifically, instead of directly training a model for brain tumor segmentation with a large amount of annotated data, we seek to train a model that can answer the question: is a voxel in the input image associated with tumor-like hyper-/hypo-intensity? Such a model can be trained by artificially generating tumor-like hyper-/hypo-intensity on images without tumors with hand-crafted designs. Since the hand-crafted designs may be too simplistic to represent all kinds of real tumors, the trained model may overfit the simplistic hand-crafted task rather than actually answer the question of abnormality. To address this problem, we propose the use of a validation task, where we generate a different hand-crafted task to monitor overfitting. In addition, we propose PL-BTS+ that further improves PL-BTS by exploiting unannotated images with brain tumors. Compared with competing unsupervised methods, the proposed method has achieved marked improvements on both public and in-house datasets, and we have also demonstrated its possible extension to other brain lesion segmentation tasks.
Knowledge Graph Completion Method Combined With Adaptive Enhanced Semantic Information
Feb 04, 2023



Abstract:Translation models tend to ignore the rich semantic information in triads in the process of knowledge graph complementation. To remedy this shortcoming, this paper constructs a knowledge graph complementation method that incorporates adaptively enhanced semantic information. The hidden semantic information inherent in the triad is obtained by fine-tuning the BERT model, and the attention feature embedding method is used to calculate the semantic attention scores between relations and entities in positive and negative triads and incorporate them into the structural information to form a soft constraint rule for semantic information. The rule is added to the original translation model to realize the adaptive enhancement of semantic information. In addition, the method takes into account the effect of high-dimensional vectors on the effect, and uses the BERT-whitening method to reduce the dimensionality and generate a more efficient semantic vector representation. After experimental comparison, the proposed method performs better on both FB15K and WIN18 datasets, with a numerical improvement of about 2.6% compared with the original translation model, which verifies the reasonableness and effectiveness of the method.
CarveMix: A Simple Data Augmentation Method for Brain Lesion Segmentation
Aug 17, 2021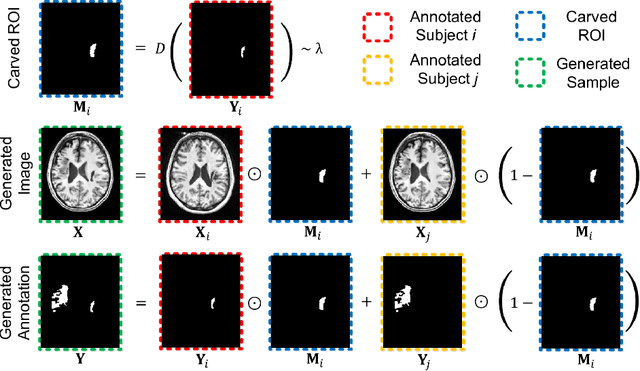
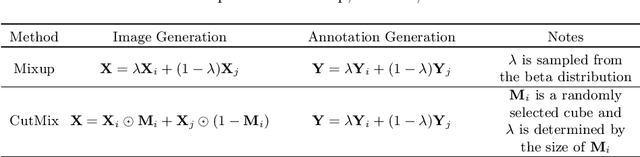
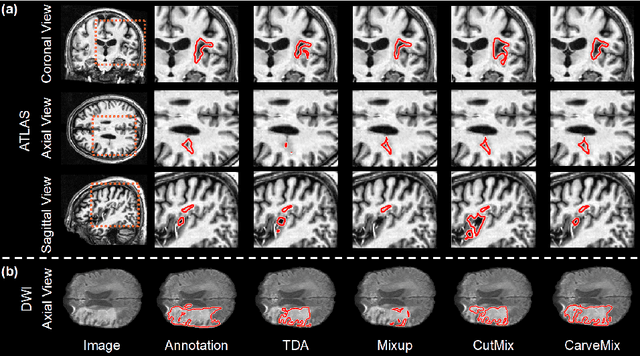
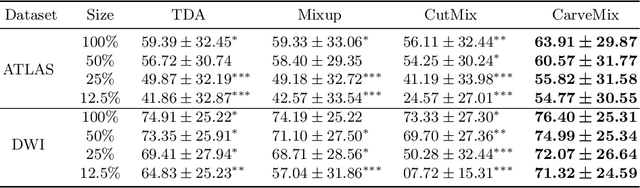
Abstract:Brain lesion segmentation provides a valuable tool for clinical diagnosis, and convolutional neural networks (CNNs) have achieved unprecedented success in the task. Data augmentation is a widely used strategy that improves the training of CNNs, and the design of the augmentation method for brain lesion segmentation is still an open problem. In this work, we propose a simple data augmentation approach, dubbed as CarveMix, for CNN-based brain lesion segmentation. Like other "mix"-based methods, such as Mixup and CutMix, CarveMix stochastically combines two existing labeled images to generate new labeled samples. Yet, unlike these augmentation strategies based on image combination, CarveMix is lesion-aware, where the combination is performed with an attention on the lesions and a proper annotation is created for the generated image. Specifically, from one labeled image we carve a region of interest (ROI) according to the lesion location and geometry, and the size of the ROI is sampled from a probability distribution. The carved ROI then replaces the corresponding voxels in a second labeled image, and the annotation of the second image is replaced accordingly as well. In this way, we generate new labeled images for network training and the lesion information is preserved. To evaluate the proposed method, experiments were performed on two brain lesion datasets. The results show that our method improves the segmentation accuracy compared with other simple data augmentation approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge