Wenjia Bai
Biomedical Image Analysis Group, Department of Computing, Imperial College London
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
Multimodal Conditional MeshGAN for Personalized Aneurysm Growth Prediction
Aug 27, 2025Abstract:Personalized, accurate prediction of aortic aneurysm progression is essential for timely intervention but remains challenging due to the need to model both subtle local deformations and global anatomical changes within complex 3D geometries. We propose MCMeshGAN, the first multimodal conditional mesh-to-mesh generative adversarial network for 3D aneurysm growth prediction. MCMeshGAN introduces a dual-branch architecture combining a novel local KNN-based convolutional network (KCN) to preserve fine-grained geometric details and a global graph convolutional network (GCN) to capture long-range structural context, overcoming the over-smoothing limitations of deep GCNs. A dedicated condition branch encodes clinical attributes (age, sex) and the target time interval to generate anatomically plausible, temporally controlled predictions, enabling retrospective and prospective modeling. We curated TAAMesh, a new longitudinal thoracic aortic aneurysm mesh dataset consisting of 590 multimodal records (CT scans, 3D meshes, and clinical data) from 208 patients. Extensive experiments demonstrate that MCMeshGAN consistently outperforms state-of-the-art baselines in both geometric accuracy and clinically important diameter estimation. This framework offers a robust step toward clinically deployable, personalized 3D disease trajectory modeling. The source code for MCMeshGAN and the baseline methods is publicly available at https://github.com/ImperialCollegeLondon/MCMeshGAN.
Knowledge to Sight: Reasoning over Visual Attributes via Knowledge Decomposition for Abnormality Grounding
Aug 06, 2025Abstract:In this work, we address the problem of grounding abnormalities in medical images, where the goal is to localize clinical findings based on textual descriptions. While generalist Vision-Language Models (VLMs) excel in natural grounding tasks, they often struggle in the medical domain due to rare, compositional, and domain-specific terms that are poorly aligned with visual patterns. Specialized medical VLMs address this challenge via large-scale domain pretraining, but at the cost of substantial annotation and computational resources. To overcome these limitations, we propose \textbf{Knowledge to Sight (K2Sight)}, a framework that introduces structured semantic supervision by decomposing clinical concepts into interpretable visual attributes, such as shape, density, and anatomical location. These attributes are distilled from domain ontologies and encoded into concise instruction-style prompts, which guide region-text alignment during training. Unlike conventional report-level supervision, our approach explicitly bridges domain knowledge and spatial structure, enabling data-efficient training of compact models. We train compact models with 0.23B and 2B parameters using only 1.5\% of the data required by state-of-the-art medical VLMs. Despite their small size and limited training data, these models achieve performance on par with or better than 7B+ medical VLMs, with up to 9.82\% improvement in $mAP_{50}$. Code and models: \href{https://lijunrio.github.io/K2Sight/}{\textcolor{SOTAPink}{https://lijunrio.github.io/K2Sight/}}.
SAM-aware Test-time Adaptation for Universal Medical Image Segmentation
Jun 05, 2025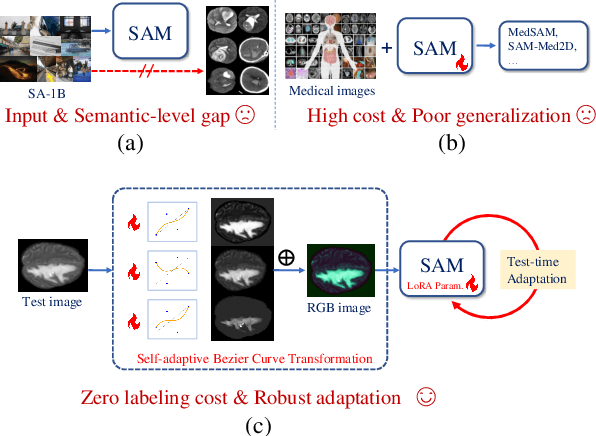
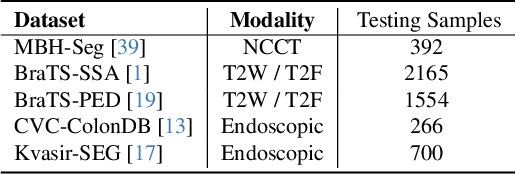
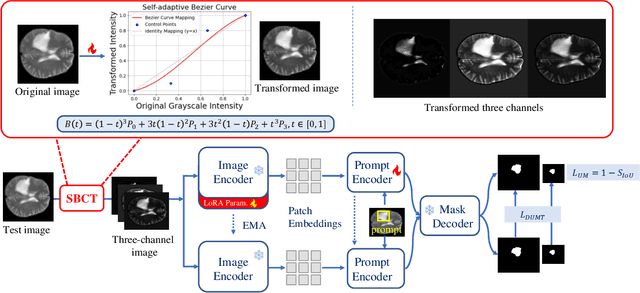
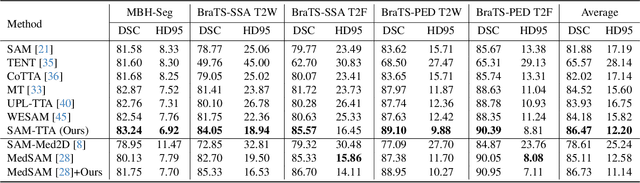
Abstract:Universal medical image segmentation using the Segment Anything Model (SAM) remains challenging due to its limited adaptability to medical domains. Existing adaptations, such as MedSAM, enhance SAM's performance in medical imaging but at the cost of reduced generalization to unseen data. Therefore, in this paper, we propose SAM-aware Test-Time Adaptation (SAM-TTA), a fundamentally different pipeline that preserves the generalization of SAM while improving its segmentation performance in medical imaging via a test-time framework. SAM-TTA tackles two key challenges: (1) input-level discrepancies caused by differences in image acquisition between natural and medical images and (2) semantic-level discrepancies due to fundamental differences in object definition between natural and medical domains (e.g., clear boundaries vs. ambiguous structures). Specifically, our SAM-TTA framework comprises (1) Self-adaptive Bezier Curve-based Transformation (SBCT), which adaptively converts single-channel medical images into three-channel SAM-compatible inputs while maintaining structural integrity, to mitigate the input gap between medical and natural images, and (2) Dual-scale Uncertainty-driven Mean Teacher adaptation (DUMT), which employs consistency learning to align SAM's internal representations to medical semantics, enabling efficient adaptation without auxiliary supervision or expensive retraining. Extensive experiments on five public datasets demonstrate that our SAM-TTA outperforms existing TTA approaches and even surpasses fully fine-tuned models such as MedSAM in certain scenarios, establishing a new paradigm for universal medical image segmentation. Code can be found at https://github.com/JianghaoWu/SAM-TTA.
Beyond Distillation: Pushing the Limits of Medical LLM Reasoning with Minimalist Rule-Based RL
May 23, 2025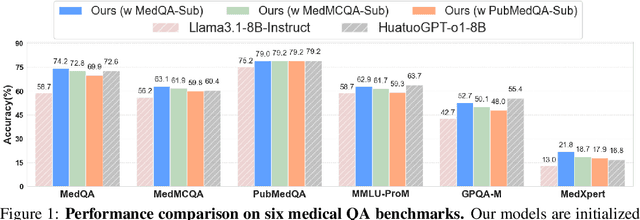
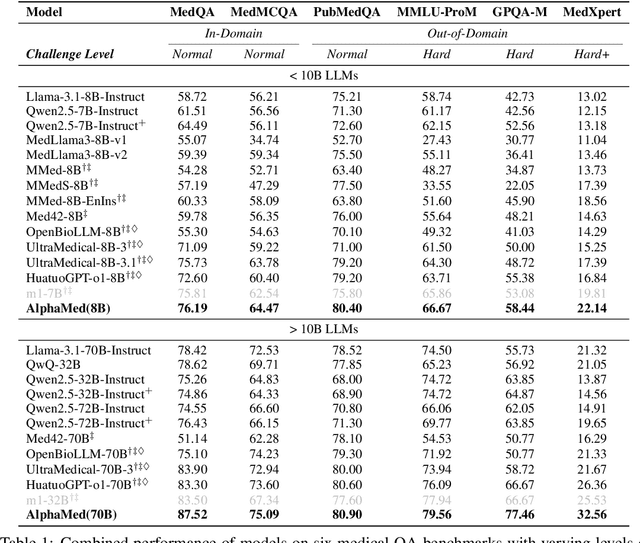
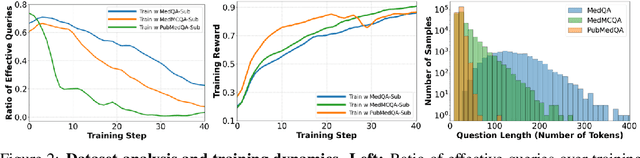
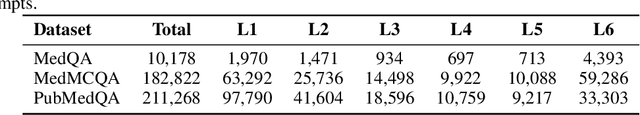
Abstract:Improving performance on complex tasks and enabling interpretable decision making in large language models (LLMs), especially for clinical applications, requires effective reasoning. Yet this remains challenging without supervised fine-tuning (SFT) on costly chain-of-thought (CoT) data distilled from closed-source models (e.g., GPT-4o). In this work, we present AlphaMed, the first medical LLM to show that reasoning capability can emerge purely through reinforcement learning (RL), using minimalist rule-based rewards on public multiple-choice QA datasets, without relying on SFT or distilled CoT data. AlphaMed achieves state-of-the-art results on six medical QA benchmarks, outperforming models trained with conventional SFT+RL pipelines. On challenging benchmarks (e.g., MedXpert), AlphaMed even surpasses larger or closed-source models such as DeepSeek-V3-671B and Claude-3.5-Sonnet. To understand the factors behind this success, we conduct a comprehensive data-centric analysis guided by three questions: (i) Can minimalist rule-based RL incentivize reasoning without distilled CoT supervision? (ii) How do dataset quantity and diversity impact reasoning? (iii) How does question difficulty shape the emergence and generalization of reasoning? Our findings show that dataset informativeness is a key driver of reasoning performance, and that minimalist RL on informative, multiple-choice QA data is effective at inducing reasoning without CoT supervision. We also observe divergent trends across benchmarks, underscoring limitations in current evaluation and the need for more challenging, reasoning-oriented medical QA benchmarks.
NOVA: A Benchmark for Anomaly Localization and Clinical Reasoning in Brain MRI
May 20, 2025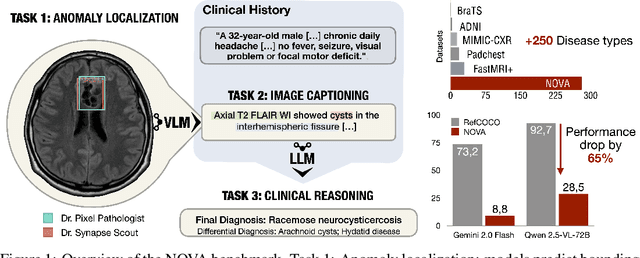

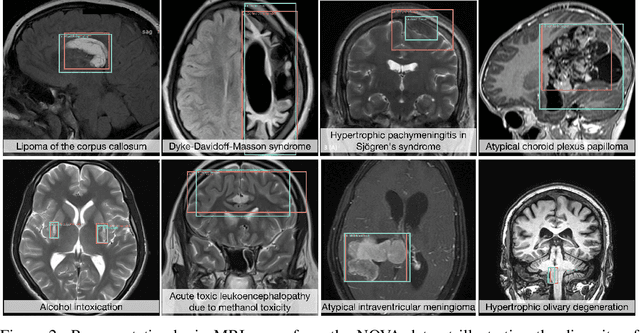
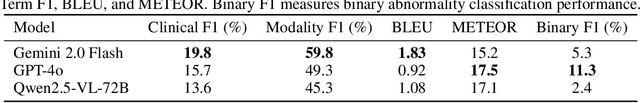
Abstract:In many real-world applications, deployed models encounter inputs that differ from the data seen during training. Out-of-distribution detection identifies whether an input stems from an unseen distribution, while open-world recognition flags such inputs to ensure the system remains robust as ever-emerging, previously $unknown$ categories appear and must be addressed without retraining. Foundation and vision-language models are pre-trained on large and diverse datasets with the expectation of broad generalization across domains, including medical imaging. However, benchmarking these models on test sets with only a few common outlier types silently collapses the evaluation back to a closed-set problem, masking failures on rare or truly novel conditions encountered in clinical use. We therefore present $NOVA$, a challenging, real-life $evaluation-only$ benchmark of $\sim$900 brain MRI scans that span 281 rare pathologies and heterogeneous acquisition protocols. Each case includes rich clinical narratives and double-blinded expert bounding-box annotations. Together, these enable joint assessment of anomaly localisation, visual captioning, and diagnostic reasoning. Because NOVA is never used for training, it serves as an $extreme$ stress-test of out-of-distribution generalisation: models must bridge a distribution gap both in sample appearance and in semantic space. Baseline results with leading vision-language models (GPT-4o, Gemini 2.0 Flash, and Qwen2.5-VL-72B) reveal substantial performance drops across all tasks, establishing NOVA as a rigorous testbed for advancing models that can detect, localize, and reason about truly unknown anomalies.
Enhancing Abnormality Grounding for Vision Language Models with Knowledge Descriptions
Mar 05, 2025Abstract:Visual Language Models (VLMs) have demonstrated impressive capabilities in visual grounding tasks. However, their effectiveness in the medical domain, particularly for abnormality detection and localization within medical images, remains underexplored. A major challenge is the complex and abstract nature of medical terminology, which makes it difficult to directly associate pathological anomaly terms with their corresponding visual features. In this work, we introduce a novel approach to enhance VLM performance in medical abnormality detection and localization by leveraging decomposed medical knowledge. Instead of directly prompting models to recognize specific abnormalities, we focus on breaking down medical concepts into fundamental attributes and common visual patterns. This strategy promotes a stronger alignment between textual descriptions and visual features, improving both the recognition and localization of abnormalities in medical images.We evaluate our method on the 0.23B Florence-2 base model and demonstrate that it achieves comparable performance in abnormality grounding to significantly larger 7B LLaVA-based medical VLMs, despite being trained on only 1.5% of the data used for such models. Experimental results also demonstrate the effectiveness of our approach in both known and previously unseen abnormalities, suggesting its strong generalization capabilities.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
Knowledge-enhanced Multimodal ECG Representation Learning with Arbitrary-Lead Inputs
Feb 25, 2025



Abstract:Recent advances in multimodal ECG representation learning center on aligning ECG signals with paired free-text reports. However, suboptimal alignment persists due to the complexity of medical language and the reliance on a full 12-lead setup, which is often unavailable in under-resourced settings. To tackle these issues, we propose **K-MERL**, a knowledge-enhanced multimodal ECG representation learning framework. **K-MERL** leverages large language models to extract structured knowledge from free-text reports and employs a lead-aware ECG encoder with dynamic lead masking to accommodate arbitrary lead inputs. Evaluations on six external ECG datasets show that **K-MERL** achieves state-of-the-art performance in zero-shot classification and linear probing tasks, while delivering an average **16%** AUC improvement over existing methods in partial-lead zero-shot classification.
SegHeD: Segmentation of Heterogeneous Data for Multiple Sclerosis Lesions with Anatomical Constraints
Oct 02, 2024Abstract:Assessment of lesions and their longitudinal progression from brain magnetic resonance (MR) images plays a crucial role in diagnosing and monitoring multiple sclerosis (MS). Machine learning models have demonstrated a great potential for automated MS lesion segmentation. Training such models typically requires large-scale high-quality datasets that are consistently annotated. However, MS imaging datasets are often small, segregated across multiple sites, with different formats (cross-sectional or longitudinal), and diverse annotation styles. This poses a significant challenge to train a unified MS lesion segmentation model. To tackle this challenge, we present SegHeD, a novel multi-dataset multi-task segmentation model that can incorporate heterogeneous data as input and perform all-lesion, new-lesion, as well as vanishing-lesion segmentation. Furthermore, we account for domain knowledge about MS lesions, incorporating longitudinal, spatial, and volumetric constraints into the segmentation model. SegHeD is assessed on five MS datasets and achieves a high performance in all, new, and vanishing-lesion segmentation, outperforming several state-of-the-art methods in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge