Fanwen Wang
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
Dynamic Dual Buffer with Divide-and-Conquer Strategy for Online Continual Learning
May 23, 2025Abstract:Online Continual Learning (OCL) presents a complex learning environment in which new data arrives in a batch-to-batch online format, and the risk of catastrophic forgetting can significantly impair model efficacy. In this study, we address OCL by introducing an innovative memory framework that incorporates a short-term memory system to retain dynamic information and a long-term memory system to archive enduring knowledge. Specifically, the long-term memory system comprises a collection of sub-memory buffers, each linked to a cluster prototype and designed to retain data samples from distinct categories. We propose a novel $K$-means-based sample selection method to identify cluster prototypes for each encountered category. To safeguard essential and critical samples, we introduce a novel memory optimisation strategy that selectively retains samples in the appropriate sub-memory buffer by evaluating each cluster prototype against incoming samples through an optimal transportation mechanism. This approach specifically promotes each sub-memory buffer to retain data samples that exhibit significant discrepancies from the corresponding cluster prototype, thereby ensuring the preservation of semantically rich information. In addition, we propose a novel Divide-and-Conquer (DAC) approach that formulates the memory updating as an optimisation problem and divides it into several subproblems. As a result, the proposed DAC approach can solve these subproblems separately and thus can significantly reduce computations of the proposed memory updating process. We conduct a series of experiments across standard and imbalanced learning settings, and the empirical findings indicate that the proposed memory framework achieves state-of-the-art performance in both learning contexts.
Enhancing Diffusion-Weighted Images (DWI) for Diffusion MRI: Is it Enough without Non-Diffusion-Weighted B=0 Reference?
May 19, 2025Abstract:Diffusion MRI (dMRI) is essential for studying brain microstructure, but high-resolution imaging remains challenging due to the inherent trade-offs between acquisition time and signal-to-noise ratio (SNR). Conventional methods often optimize only the diffusion-weighted images (DWIs) without considering their relationship with the non-diffusion-weighted (b=0) reference images. However, calculating diffusion metrics, such as the apparent diffusion coefficient (ADC) and diffusion tensor with its derived metrics like fractional anisotropy (FA) and mean diffusivity (MD), relies on the ratio between each DWI and the b=0 image, which is crucial for clinical observation and diagnostics. In this study, we demonstrate that solely enhancing DWIs using a conventional pixel-wise mean squared error (MSE) loss is insufficient, as the error in ratio between generated DWIs and b=0 diverges. We propose a novel ratio loss, defined as the MSE loss between the predicted and ground-truth log of DWI/b=0 ratios. Our results show that incorporating the ratio loss significantly improves the convergence of this ratio error, achieving lower ratio MSE and slightly enhancing the peak signal-to-noise ratio (PSNR) of generated DWIs. This leads to improved dMRI super-resolution and better preservation of b=0 ratio-based features for the derivation of diffusion metrics.
Decoupling Multi-Contrast Super-Resolution: Pairing Unpaired Synthesis with Implicit Representations
May 09, 2025Abstract:Magnetic Resonance Imaging (MRI) is critical for clinical diagnostics but is often limited by long acquisition times and low signal-to-noise ratios, especially in modalities like diffusion and functional MRI. The multi-contrast nature of MRI presents a valuable opportunity for cross-modal enhancement, where high-resolution (HR) modalities can serve as references to boost the quality of their low-resolution (LR) counterparts-motivating the development of Multi-Contrast Super-Resolution (MCSR) techniques. Prior work has shown that leveraging complementary contrasts can improve SR performance; however, effective feature extraction and fusion across modalities with varying resolutions remains a major challenge. Moreover, existing MCSR methods often assume fixed resolution settings and all require large, perfectly paired training datasets-conditions rarely met in real-world clinical environments. To address these challenges, we propose a novel Modular Multi-Contrast Super-Resolution (MCSR) framework that eliminates the need for paired training data and supports arbitrary upscaling. Our method decouples the MCSR task into two stages: (1) Unpaired Cross-Modal Synthesis (U-CMS), which translates a high-resolution reference modality into a synthesized version of the target contrast, and (2) Unsupervised Super-Resolution (U-SR), which reconstructs the final output using implicit neural representations (INRs) conditioned on spatial coordinates. This design enables scale-agnostic and anatomically faithful reconstruction by bridging un-paired cross-modal synthesis with unsupervised resolution enhancement. Experiments show that our method achieves superior performance at 4x and 8x upscaling, with improved fidelity and anatomical consistency over existing baselines. Our framework demonstrates strong potential for scalable, subject-specific, and data-efficient MCSR in real-world clinical settings.
RSFR: A Coarse-to-Fine Reconstruction Framework for Diffusion Tensor Cardiac MRI with Semantic-Aware Refinement
Apr 25, 2025Abstract:Cardiac diffusion tensor imaging (DTI) offers unique insights into cardiomyocyte arrangements, bridging the gap between microscopic and macroscopic cardiac function. However, its clinical utility is limited by technical challenges, including a low signal-to-noise ratio, aliasing artefacts, and the need for accurate quantitative fidelity. To address these limitations, we introduce RSFR (Reconstruction, Segmentation, Fusion & Refinement), a novel framework for cardiac diffusion-weighted image reconstruction. RSFR employs a coarse-to-fine strategy, leveraging zero-shot semantic priors via the Segment Anything Model and a robust Vision Mamba-based reconstruction backbone. Our framework integrates semantic features effectively to mitigate artefacts and enhance fidelity, achieving state-of-the-art reconstruction quality and accurate DT parameter estimation under high undersampling rates. Extensive experiments and ablation studies demonstrate the superior performance of RSFR compared to existing methods, highlighting its robustness, scalability, and potential for clinical translation in quantitative cardiac DTI.
Is fitting error a reliable metric for assessing deformable motion correction in quantitative MRI?
Mar 10, 2025Abstract:Quantitative MR (qMR) can provide numerical values representing the physical and chemical properties of the tissues. To collect a series of frames under varying settings, retrospective motion correction is essential to align the corresponding anatomical points or features. Under the assumption that the misalignment makes the discrepancy between the corresponding features larger, fitting error is a commonly used evaluation metric for motion correction in qMR. This study evaluates the reliability of the fitting error metric in cardiac diffusion tensor imaging (cDTI) after deformable registration. We found that while fitting error correlates with the negative eigenvalues, the negative Jacobian Determinant increases with broken cardiomyocytes, indicated by helix angle gradient line profiles. Since fitting error measures the distance between moved points and their re-rendered counterparts, the fitting parameter itself may be adjusted due to poor registration. Therefore, fitting error in deformable registration itself is a necessary but not sufficient metric and should be combined with other metrics.
Lightweight Hypercomplex MRI Reconstruction: A Generalized Kronecker-Parameterized Approach
Mar 07, 2025Abstract:Magnetic Resonance Imaging (MRI) is crucial for clinical diagnostics but is hindered by prolonged scan times. Current deep learning models enhance MRI reconstruction but are often memory-intensive and unsuitable for resource-limited systems. This paper introduces a lightweight MRI reconstruction model leveraging Kronecker-Parameterized Hypercomplex Neural Networks to achieve high performance with reduced parameters. By integrating Kronecker-based modules, including Kronecker MLP, Kronecker Window Attention, and Kronecker Convolution, the proposed model efficiently extracts spatial features while preserving representational power. We introduce Kronecker U-Net and Kronecker SwinMR, which maintain high reconstruction quality with approximately 50% fewer parameters compared to existing models. Experimental evaluation on the FastMRI dataset demonstrates competitive PSNR, SSIM, and LPIPS metrics, even at high acceleration factors (8x and 16x), with no significant performance drop. Additionally, Kronecker variants exhibit superior generalization and reduced overfitting on limited datasets, facilitating efficient MRI reconstruction on hardware-constrained systems. This approach sets a new benchmark for parameter-efficient medical imaging models.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
Explicit Differentiable Slicing and Global Deformation for Cardiac Mesh Reconstruction
Sep 03, 2024



Abstract:Mesh reconstruction of the cardiac anatomy from medical images is useful for shape and motion measurements and biophysics simulations to facilitate the assessment of cardiac function and health. However, 3D medical images are often acquired as 2D slices that are sparsely sampled and noisy, and mesh reconstruction on such data is a challenging task. Traditional voxel-based approaches rely on pre- and post-processing that compromises image fidelity, while mesh-level deep learning approaches require mesh annotations that are difficult to get. Therefore, direct cross-domain supervision from 2D images to meshes is a key technique for advancing 3D learning in medical imaging, but it has not been well-developed. While there have been attempts to approximate the optimized meshes' slicing, few existing methods directly use 2D slices to supervise mesh reconstruction in a differentiable manner. Here, we propose a novel explicit differentiable voxelization and slicing (DVS) algorithm that allows gradient backpropagation to a mesh from its slices, facilitating refined mesh optimization directly supervised by the losses defined on 2D images. Further, we propose an innovative framework for extracting patient-specific left ventricle (LV) meshes from medical images by coupling DVS with a graph harmonic deformation (GHD) mesh morphing descriptor of cardiac shape that naturally preserves mesh quality and smoothness during optimization. Experimental results demonstrate that our method achieves state-of-the-art performance in cardiac mesh reconstruction tasks from CT and MRI, with an overall Dice score of 90% on multi-datasets, outperforming existing approaches. The proposed method can further quantify clinically useful parameters such as ejection fraction and global myocardial strains, closely matching the ground truth and surpassing the traditional voxel-based approach in sparse images.
CMRxRecon2024: A Multi-Modality, Multi-View K-Space Dataset Boosting Universal Machine Learning for Accelerated Cardiac MRI
Jun 27, 2024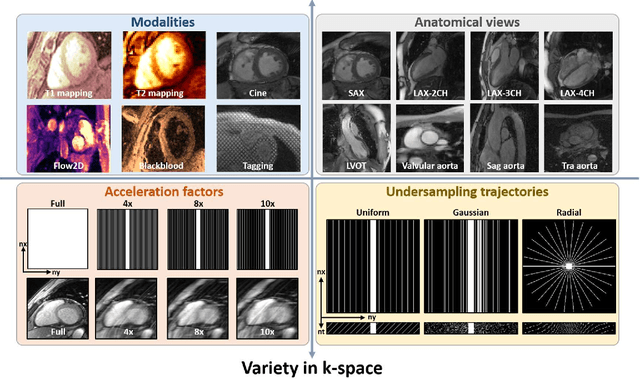
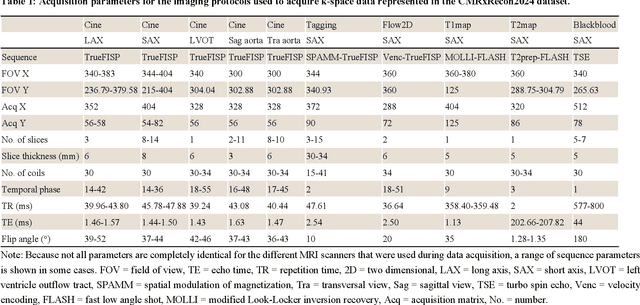
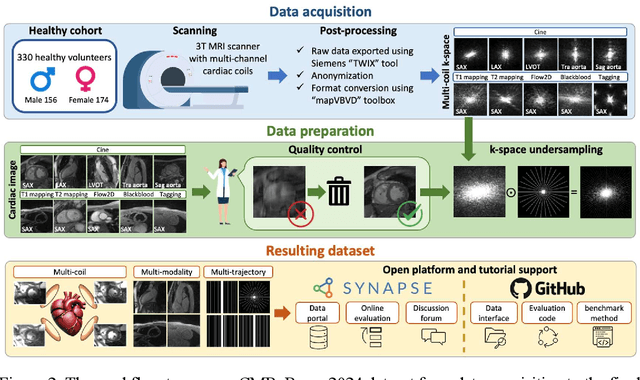
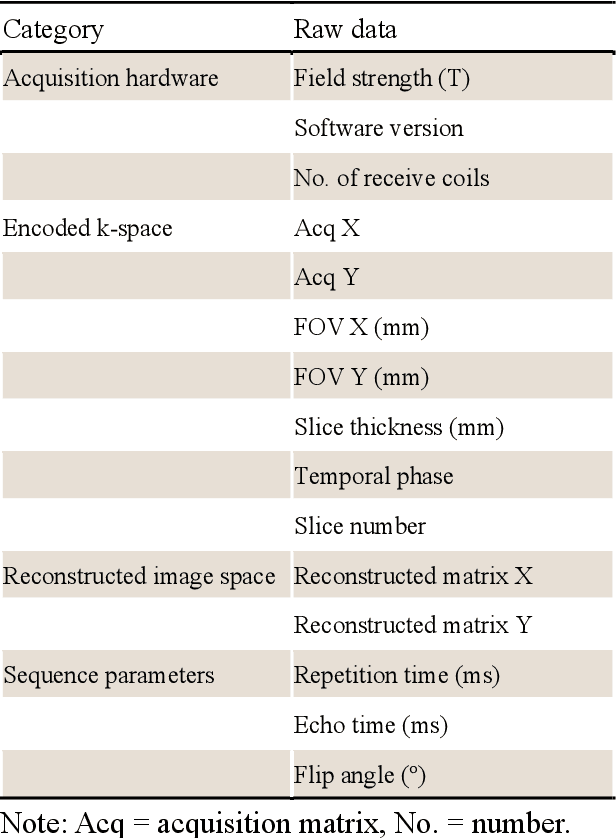
Abstract:Cardiac magnetic resonance imaging (MRI) has emerged as a clinically gold-standard technique for diagnosing cardiac diseases, thanks to its ability to provide diverse information with multiple modalities and anatomical views. Accelerated cardiac MRI is highly expected to achieve time-efficient and patient-friendly imaging, and then advanced image reconstruction approaches are required to recover high-quality, clinically interpretable images from undersampled measurements. However, the lack of publicly available cardiac MRI k-space dataset in terms of both quantity and diversity has severely hindered substantial technological progress, particularly for data-driven artificial intelligence. Here, we provide a standardized, diverse, and high-quality CMRxRecon2024 dataset to facilitate the technical development, fair evaluation, and clinical transfer of cardiac MRI reconstruction approaches, towards promoting the universal frameworks that enable fast and robust reconstructions across different cardiac MRI protocols in clinical practice. To the best of our knowledge, the CMRxRecon2024 dataset is the largest and most diverse publicly available cardiac k-space dataset. It is acquired from 330 healthy volunteers, covering commonly used modalities, anatomical views, and acquisition trajectories in clinical cardiac MRI workflows. Besides, an open platform with tutorials, benchmarks, and data processing tools is provided to facilitate data usage, advanced method development, and fair performance evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge