Kunyuan Guo
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
CMRxRecon2024: A Multi-Modality, Multi-View K-Space Dataset Boosting Universal Machine Learning for Accelerated Cardiac MRI
Jun 27, 2024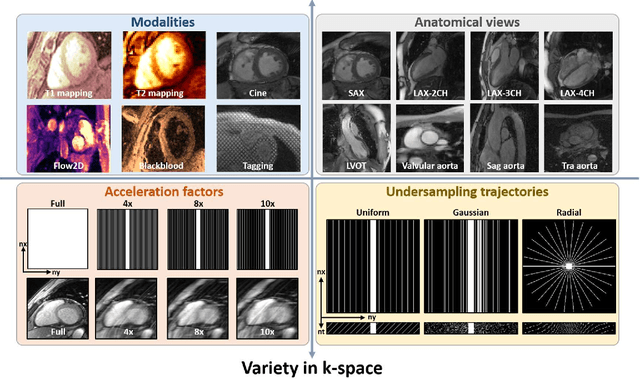
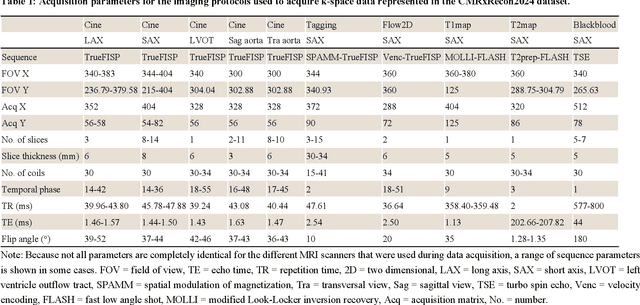
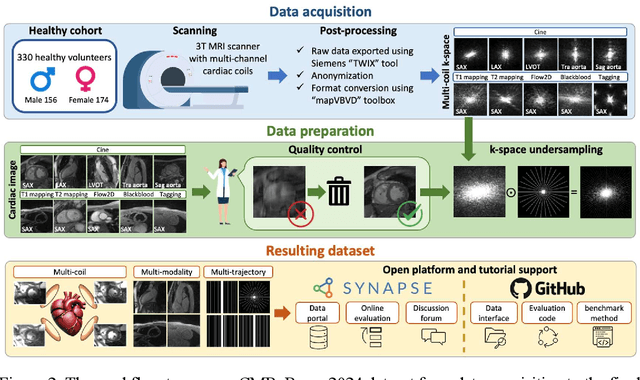
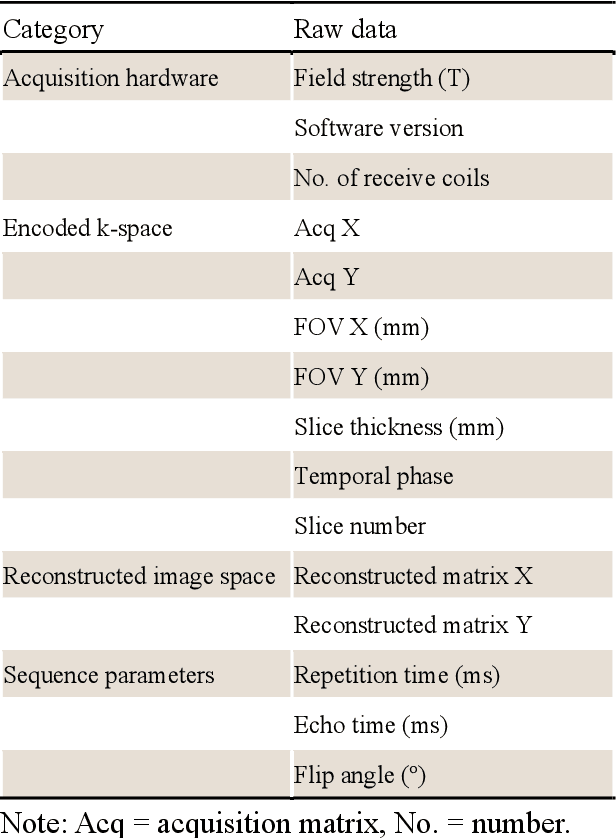
Abstract:Cardiac magnetic resonance imaging (MRI) has emerged as a clinically gold-standard technique for diagnosing cardiac diseases, thanks to its ability to provide diverse information with multiple modalities and anatomical views. Accelerated cardiac MRI is highly expected to achieve time-efficient and patient-friendly imaging, and then advanced image reconstruction approaches are required to recover high-quality, clinically interpretable images from undersampled measurements. However, the lack of publicly available cardiac MRI k-space dataset in terms of both quantity and diversity has severely hindered substantial technological progress, particularly for data-driven artificial intelligence. Here, we provide a standardized, diverse, and high-quality CMRxRecon2024 dataset to facilitate the technical development, fair evaluation, and clinical transfer of cardiac MRI reconstruction approaches, towards promoting the universal frameworks that enable fast and robust reconstructions across different cardiac MRI protocols in clinical practice. To the best of our knowledge, the CMRxRecon2024 dataset is the largest and most diverse publicly available cardiac k-space dataset. It is acquired from 330 healthy volunteers, covering commonly used modalities, anatomical views, and acquisition trajectories in clinical cardiac MRI workflows. Besides, an open platform with tutorials, benchmarks, and data processing tools is provided to facilitate data usage, advanced method development, and fair performance evaluation.
Simultaneous Deep Learning of Myocardium Segmentation and T2 Quantification for Acute Myocardial Infarction MRI
May 17, 2024



Abstract:In cardiac Magnetic Resonance Imaging (MRI) analysis, simultaneous myocardial segmentation and T2 quantification are crucial for assessing myocardial pathologies. Existing methods often address these tasks separately, limiting their synergistic potential. To address this, we propose SQNet, a dual-task network integrating Transformer and Convolutional Neural Network (CNN) components. SQNet features a T2-refine fusion decoder for quantitative analysis, leveraging global features from the Transformer, and a segmentation decoder with multiple local region supervision for enhanced accuracy. A tight coupling module aligns and fuses CNN and Transformer branch features, enabling SQNet to focus on myocardium regions. Evaluation on healthy controls (HC) and acute myocardial infarction patients (AMI) demonstrates superior segmentation dice scores (89.3/89.2) compared to state-of-the-art methods (87.7/87.9). T2 quantification yields strong linear correlations (Pearson coefficients: 0.84/0.93) with label values for HC/AMI, indicating accurate mapping. Radiologist evaluations confirm SQNet's superior image quality scores (4.60/4.58 for segmentation, 4.32/4.42 for T2 quantification) over state-of-the-art methods (4.50/4.44 for segmentation, 3.59/4.37 for T2 quantification). SQNet thus offers accurate simultaneous segmentation and quantification, enhancing cardiac disease diagnosis, such as AMI.
The state-of-the-art in Cardiac MRI Reconstruction: Results of the CMRxRecon Challenge in MICCAI 2023
Apr 01, 2024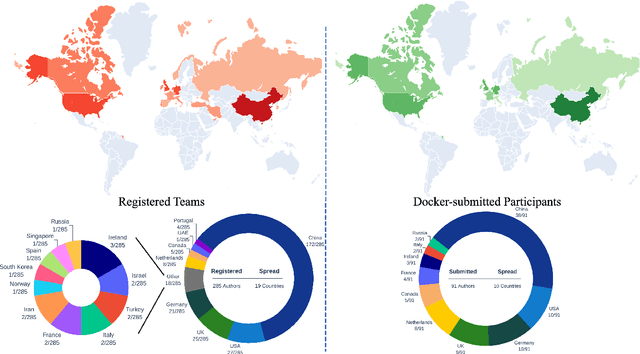
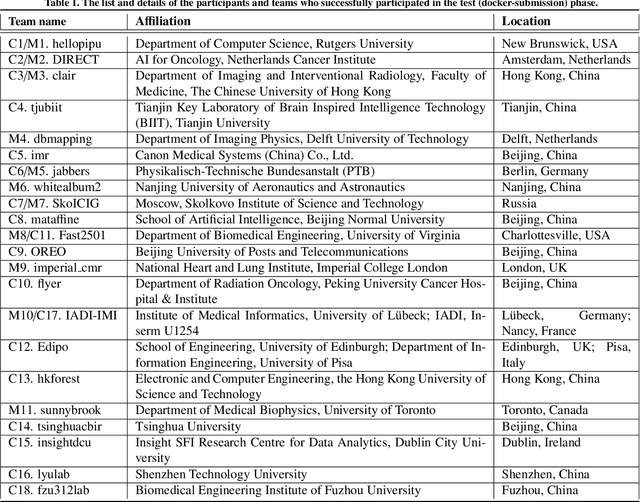
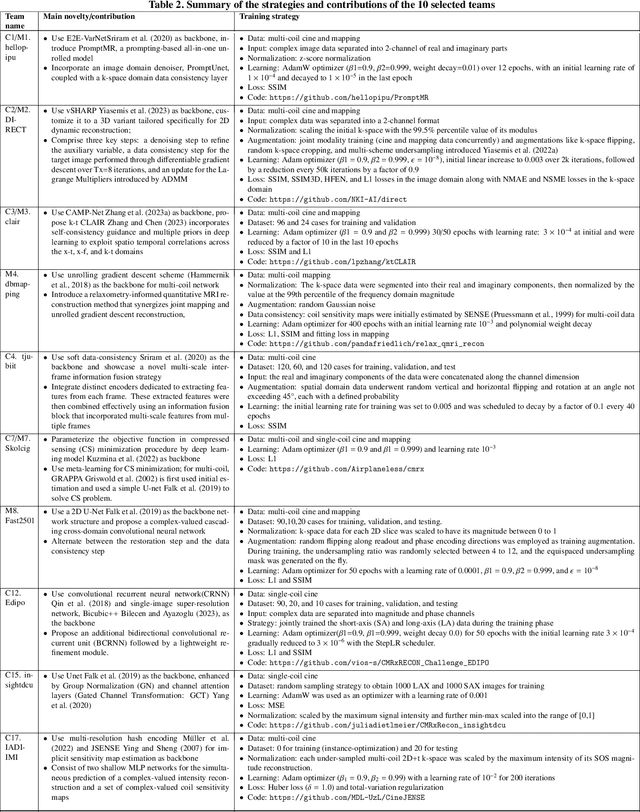
Abstract:Cardiac MRI, crucial for evaluating heart structure and function, faces limitations like slow imaging and motion artifacts. Undersampling reconstruction, especially data-driven algorithms, has emerged as a promising solution to accelerate scans and enhance imaging performance using highly under-sampled data. Nevertheless, the scarcity of publicly available cardiac k-space datasets and evaluation platform hinder the development of data-driven reconstruction algorithms. To address this issue, we organized the Cardiac MRI Reconstruction Challenge (CMRxRecon) in 2023, in collaboration with the 26th International Conference on MICCAI. CMRxRecon presented an extensive k-space dataset comprising cine and mapping raw data, accompanied by detailed annotations of cardiac anatomical structures. With overwhelming participation, the challenge attracted more than 285 teams and over 600 participants. Among them, 22 teams successfully submitted Docker containers for the testing phase, with 7 teams submitted for both cine and mapping tasks. All teams use deep learning based approaches, indicating that deep learning has predominately become a promising solution for the problem. The first-place winner of both tasks utilizes the E2E-VarNet architecture as backbones. In contrast, U-Net is still the most popular backbone for both multi-coil and single-coil reconstructions. This paper provides a comprehensive overview of the challenge design, presents a summary of the submitted results, reviews the employed methods, and offers an in-depth discussion that aims to inspire future advancements in cardiac MRI reconstruction models. The summary emphasizes the effective strategies observed in Cardiac MRI reconstruction, including backbone architecture, loss function, pre-processing techniques, physical modeling, and model complexity, thereby providing valuable insights for further developments in this field.
CMRxRecon: An open cardiac MRI dataset for the competition of accelerated image reconstruction
Sep 19, 2023



Abstract:Cardiac magnetic resonance imaging (CMR) has emerged as a valuable diagnostic tool for cardiac diseases. However, a limitation of CMR is its slow imaging speed, which causes patient discomfort and introduces artifacts in the images. There has been growing interest in deep learning-based CMR imaging algorithms that can reconstruct high-quality images from highly under-sampled k-space data. However, the development of deep learning methods requires large training datasets, which have not been publicly available for CMR. To address this gap, we released a dataset that includes multi-contrast, multi-view, multi-slice and multi-coil CMR imaging data from 300 subjects. Imaging studies include cardiac cine and mapping sequences. Manual segmentations of the myocardium and chambers of all the subjects are also provided within the dataset. Scripts of state-of-the-art reconstruction algorithms were also provided as a point of reference. Our aim is to facilitate the advancement of state-of-the-art CMR image reconstruction by introducing standardized evaluation criteria and making the dataset freely accessible to the research community. Researchers can access the dataset at https://www.synapse.org/#!Synapse:syn51471091/wiki/.
Physics-driven Synthetic Data Learning for Biomedical Magnetic Resonance
Mar 22, 2022



Abstract:Deep learning has innovated the field of computational imaging. One of its bottlenecks is unavailable or insufficient training data. This article reviews an emerging paradigm, imaging physics-based data synthesis (IPADS), that can provide huge training data in biomedical magnetic resonance without or with few real data. Following the physical law of magnetic resonance, IPADS generates signals from differential equations or analytical solution models, making the learning more scalable, explainable, and better protecting privacy. Key components of IPADS learning, including signal generation models, basic deep learning network structures, enhanced data generation, and learning methods are discussed. Great potentials of IPADS have been demonstrated by representative applications in fast imaging, ultrafast signal reconstruction and accurate parameter quantification. Finally, open questions and future work have been discussed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge