Chen Qin
Self-Supervised Slice-to-Volume Reconstruction with Gaussian Representations for Fetal MRI
Jan 30, 2026Abstract:Reconstructing 3D fetal MR volumes from motion-corrupted stacks of 2D slices is a crucial and challenging task. Conventional slice-to-volume reconstruction (SVR) methods are time-consuming and require multiple orthogonal stacks for reconstruction. While learning-based SVR approaches have significantly reduced the time required at the inference stage, they heavily rely on ground truth information for training, which is inaccessible in practice. To address these challenges, we propose GaussianSVR, a self-supervised framework for slice-to-volume reconstruction. GaussianSVR represents the target volume using 3D Gaussian representations to achieve high-fidelity reconstruction. It leverages a simulated forward slice acquisition model to enable self-supervised training, alleviating the need for ground-truth volumes. Furthermore, to enhance both accuracy and efficiency, we introduce a multi-resolution training strategy that jointly optimizes Gaussian parameters and spatial transformations across different resolution levels. Experiments show that GaussianSVR outperforms the baseline methods on fetal MR volumetric reconstruction. Code will be available upon acceptance.
Inference-Time Dynamic Modality Selection for Incomplete Multimodal Classification
Jan 30, 2026Abstract:Multimodal deep learning (MDL) has achieved remarkable success across various domains, yet its practical deployment is often hindered by incomplete multimodal data. Existing incomplete MDL methods either discard missing modalities, risking the loss of valuable task-relevant information, or recover them, potentially introducing irrelevant noise, leading to the discarding-imputation dilemma. To address this dilemma, in this paper, we propose DyMo, a new inference-time dynamic modality selection framework that adaptively identifies and integrates reliable recovered modalities, fully exploring task-relevant information beyond the conventional discard-or-impute paradigm. Central to DyMo is a novel selection algorithm that maximizes multimodal task-relevant information for each test sample. Since direct estimation of such information at test time is intractable due to the unknown data distribution, we theoretically establish a connection between information and the task loss, which we compute at inference time as a tractable proxy. Building on this, a novel principled reward function is proposed to guide modality selection. In addition, we design a flexible multimodal network architecture compatible with arbitrary modality combinations, alongside a tailored training strategy for robust representation learning. Extensive experiments on diverse natural and medical image datasets show that DyMo significantly outperforms state-of-the-art incomplete/dynamic MDL methods across various missing-data scenarios. Our code is available at https://github.com//siyi-wind/DyMo.
Adaptive Conditional Contrast-Agnostic Deformable Image Registration with Uncertainty Estimation
Jan 09, 2026Abstract:Deformable multi-contrast image registration is a challenging yet crucial task due to the complex, non-linear intensity relationships across different imaging contrasts. Conventional registration methods typically rely on iterative optimization of the deformation field, which is time-consuming. Although recent learning-based approaches enable fast and accurate registration during inference, their generalizability remains limited to the specific contrasts observed during training. In this work, we propose an adaptive conditional contrast-agnostic deformable image registration framework (AC-CAR) based on a random convolution-based contrast augmentation scheme. AC-CAR can generalize to arbitrary imaging contrasts without observing them during training. To encourage contrast-invariant feature learning, we propose an adaptive conditional feature modulator (ACFM) that adaptively modulates the features and the contrast-invariant latent regularization to enforce the consistency of the learned feature across different imaging contrasts. Additionally, we enable our framework to provide contrast-agnostic registration uncertainty by integrating a variance network that leverages the contrast-agnostic registration encoder to improve the trustworthiness and reliability of AC-CAR. Experimental results demonstrate that AC-CAR outperforms baseline methods in registration accuracy and exhibits superior generalization to unseen imaging contrasts. Code is available at https://github.com/Yinsong0510/AC-CAR.
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
UPMRI: Unsupervised Parallel MRI Reconstruction via Projected Conditional Flow Matching
Dec 19, 2025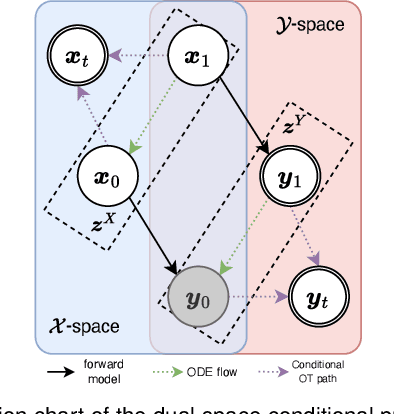
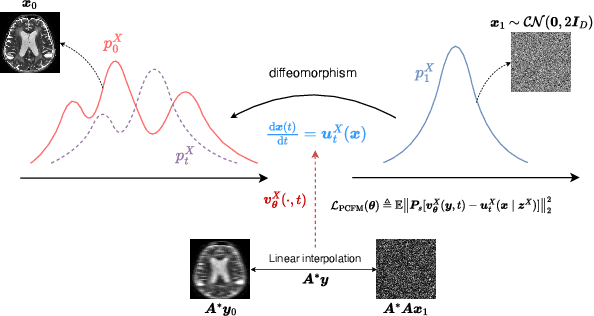
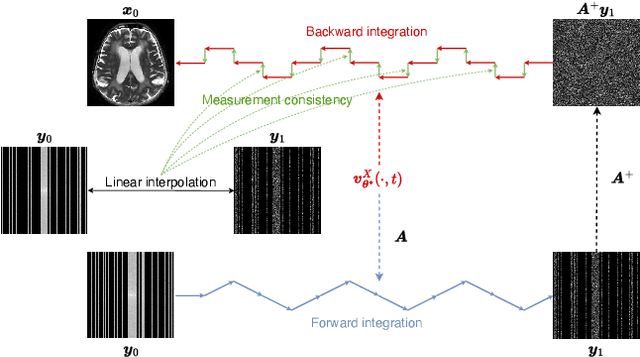
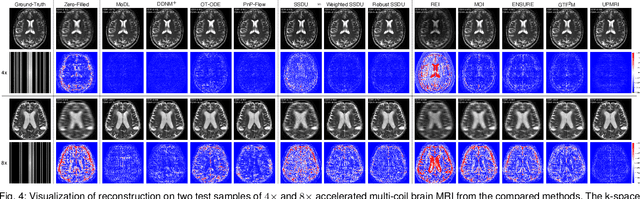
Abstract:Reconstructing high-quality images from substantially undersampled k-space data for accelerated MRI presents a challenging ill-posed inverse problem. While supervised deep learning has revolutionized this field, it relies heavily on large datasets of fully sampled ground-truth images, which are often impractical or impossible to acquire in clinical settings due to long scan times. Despite advances in self-supervised/unsupervised MRI reconstruction, their performance remains inadequate at high acceleration rates. To bridge this gap, we introduce UPMRI, an unsupervised reconstruction framework based on Projected Conditional Flow Matching (PCFM) and its unsupervised transformation. Unlike standard generative models, PCFM learns the prior distribution of fully sampled parallel MRI data by utilizing only undersampled k-space measurements. To reconstruct the image, we establish a novel theoretical link between the marginal vector field in the measurement space, governed by the continuity equation, and the optimal solution to the PCFM objective. This connection results in a cyclic dual-space sampling algorithm for high-quality reconstruction. Extensive evaluations on the fastMRI brain and CMRxRecon cardiac datasets demonstrate that UPMRI significantly outperforms state-of-the-art self-supervised and unsupervised baselines. Notably, it also achieves reconstruction fidelity comparable to or better than leading supervised methods at high acceleration factors, while requiring no fully sampled training data.
Spectral Bias Correction in PINNs for Myocardial Image Registration of Pathological Data
Apr 24, 2025



Abstract:Accurate myocardial image registration is essential for cardiac strain analysis and disease diagnosis. However, spectral bias in neural networks impedes modeling high-frequency deformations, producing inaccurate, biomechanically implausible results, particularly in pathological data. This paper addresses spectral bias in physics-informed neural networks (PINNs) by integrating Fourier Feature mappings and introducing modulation strategies into a PINN framework. Experiments on two distinct datasets demonstrate that the proposed methods enhance the PINN's ability to capture complex, high-frequency deformations in cardiomyopathies, achieving superior registration accuracy while maintaining biomechanical plausibility - thus providing a foundation for scalable cardiac image registration and generalization across multiple patients and pathologies.
STiL: Semi-supervised Tabular-Image Learning for Comprehensive Task-Relevant Information Exploration in Multimodal Classification
Mar 08, 2025Abstract:Multimodal image-tabular learning is gaining attention, yet it faces challenges due to limited labeled data. While earlier work has applied self-supervised learning (SSL) to unlabeled data, its task-agnostic nature often results in learning suboptimal features for downstream tasks. Semi-supervised learning (SemiSL), which combines labeled and unlabeled data, offers a promising solution. However, existing multimodal SemiSL methods typically focus on unimodal or modality-shared features, ignoring valuable task-relevant modality-specific information, leading to a Modality Information Gap. In this paper, we propose STiL, a novel SemiSL tabular-image framework that addresses this gap by comprehensively exploring task-relevant information. STiL features a new disentangled contrastive consistency module to learn cross-modal invariant representations of shared information while retaining modality-specific information via disentanglement. We also propose a novel consensus-guided pseudo-labeling strategy to generate reliable pseudo-labels based on classifier consensus, along with a new prototype-guided label smoothing technique to refine pseudo-label quality with prototype embeddings, thereby enhancing task-relevant information learning in unlabeled data. Experiments on natural and medical image datasets show that STiL outperforms the state-of-the-art supervised/SSL/SemiSL image/multimodal approaches. Our code is publicly available.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
Unsupervised Accelerated MRI Reconstruction via Ground-Truth-Free Flow Matching
Feb 24, 2025Abstract:Accelerated magnetic resonance imaging involves reconstructing fully sampled images from undersampled k-space measurements. Current state-of-the-art approaches have mainly focused on either end-to-end supervised training inspired by compressed sensing formulations, or posterior sampling methods built on modern generative models. However, their efficacy heavily relies on large datasets of fully sampled images, which may not always be available in practice. To address this issue, we propose an unsupervised MRI reconstruction method based on ground-truth-free flow matching (GTF$^2$M). Particularly, the GTF$^2$M learns a prior denoising process of fully sampled ground-truth images using only undersampled data. Based on that, an efficient cyclic reconstruction algorithm is further proposed to perform forward and backward integration in the dual space of image-space signal and k-space measurement. We compared our method with state-of-the-art learning-based baselines on the fastMRI database of both single-coil knee and multi-coil brain MRIs. The results show that our proposed unsupervised method can significantly outperform existing unsupervised approaches, and achieve performance comparable to most supervised end-to-end and prior learning baselines trained on fully sampled MRI, while offering greater efficiency than the compared generative model-based approaches.
Uncertainty quantification for White Matter Hyperintensity segmentation detects silent failures and improves automated Fazekas quantification
Nov 26, 2024Abstract:White Matter Hyperintensities (WMH) are key neuroradiological markers of small vessel disease present in brain MRI. Assessment of WMH is important in research and clinics. However, WMH are challenging to segment due to their high variability in shape, location, size, poorly defined borders, and similar intensity profile to other pathologies (e.g stroke lesions) and artefacts (e.g head motion). In this work, we apply the most effective techniques for uncertainty quantification (UQ) in segmentation to the WMH segmentation task across multiple test-time data distributions. We find a combination of Stochastic Segmentation Networks with Deep Ensembles yields the highest Dice and lowest Absolute Volume Difference % (AVD) score on in-domain and out-of-distribution data. We demonstrate the downstream utility of UQ, proposing a novel method for classification of the clinical Fazekas score using spatial features extracted for WMH segmentation and UQ maps. We show that incorporating WMH uncertainty information improves Fazekas classification performance and calibration, with median class balanced accuracy for classification models with (UQ and spatial WMH features)/(spatial WMH features)/(WMH volume only) of 0.71/0.66/0.60 in the Deep WMH and 0.82/0.77/0.73 in the Periventricular WMH regions respectively. We demonstrate that stochastic UQ techniques with high sample diversity can improve the detection of poor quality segmentations. Finally, we qualitatively analyse the semantic information captured by UQ techniques and demonstrate that uncertainty can highlight areas where there is ambiguity between WMH and stroke lesions, while identifying clusters of small WMH in deep white matter unsegmented by the model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge