Ben Philps
Uncertainty quantification for White Matter Hyperintensity segmentation detects silent failures and improves automated Fazekas quantification
Nov 26, 2024Abstract:White Matter Hyperintensities (WMH) are key neuroradiological markers of small vessel disease present in brain MRI. Assessment of WMH is important in research and clinics. However, WMH are challenging to segment due to their high variability in shape, location, size, poorly defined borders, and similar intensity profile to other pathologies (e.g stroke lesions) and artefacts (e.g head motion). In this work, we apply the most effective techniques for uncertainty quantification (UQ) in segmentation to the WMH segmentation task across multiple test-time data distributions. We find a combination of Stochastic Segmentation Networks with Deep Ensembles yields the highest Dice and lowest Absolute Volume Difference % (AVD) score on in-domain and out-of-distribution data. We demonstrate the downstream utility of UQ, proposing a novel method for classification of the clinical Fazekas score using spatial features extracted for WMH segmentation and UQ maps. We show that incorporating WMH uncertainty information improves Fazekas classification performance and calibration, with median class balanced accuracy for classification models with (UQ and spatial WMH features)/(spatial WMH features)/(WMH volume only) of 0.71/0.66/0.60 in the Deep WMH and 0.82/0.77/0.73 in the Periventricular WMH regions respectively. We demonstrate that stochastic UQ techniques with high sample diversity can improve the detection of poor quality segmentations. Finally, we qualitatively analyse the semantic information captured by UQ techniques and demonstrate that uncertainty can highlight areas where there is ambiguity between WMH and stroke lesions, while identifying clusters of small WMH in deep white matter unsegmented by the model.
InstanSeg: an embedding-based instance segmentation algorithm optimized for accurate, efficient and portable cell segmentation
Aug 28, 2024
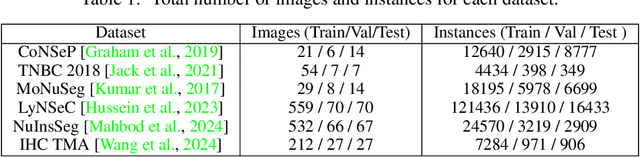

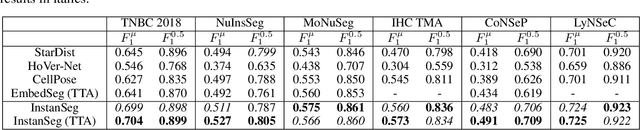
Abstract:Cell and nucleus segmentation are fundamental tasks for quantitative bioimage analysis. Despite progress in recent years, biologists and other domain experts still require novel algorithms to handle increasingly large and complex real-world datasets. These algorithms must not only achieve state-of-the-art accuracy, but also be optimized for efficiency, portability and user-friendliness. Here, we introduce InstanSeg: a novel embedding-based instance segmentation pipeline designed to identify cells and nuclei in microscopy images. Using six public cell segmentation datasets, we demonstrate that InstanSeg can significantly improve accuracy when compared to the most widely used alternative methods, while reducing the processing time by at least 60%. Furthermore, InstanSeg is designed to be fully serializable as TorchScript and supports GPU acceleration on a range of hardware. We provide an open-source implementation of InstanSeg in Python, in addition to a user-friendly, interactive QuPath extension for inference written in Java. Our code and pre-trained models are available at https://github.com/instanseg/instanseg .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge