Joanna M. Wardlaw
Comparative evaluation of training strategies using partially labelled datasets for segmentation of white matter hyperintensities and stroke lesions in FLAIR MRI
Jan 28, 2026Abstract:White matter hyperintensities (WMH) and ischaemic stroke lesions (ISL) are imaging features associated with cerebral small vessel disease (SVD) that are visible on brain magnetic resonance imaging (MRI) scans. The development and validation of deep learning models to segment and differentiate these features is difficult because they visually confound each other in the fluid-attenuated inversion recovery (FLAIR) sequence and often appear in the same subject. We investigated six strategies for training a combined WMH and ISL segmentation model using partially labelled data. We combined privately held fully and partially labelled datasets with publicly available partially labelled datasets to yield a total of 2052 MRI volumes, with 1341 and 1152 containing ground truth annotations for WMH and ISL respectively. We found that several methods were able to effectively leverage the partially labelled data to improve model performance, with the use of pseudolabels yielding the best result.
Standardized Evaluation of Automatic Methods for Perivascular Spaces Segmentation in MRI -- MICCAI 2024 Challenge Results
Dec 20, 2025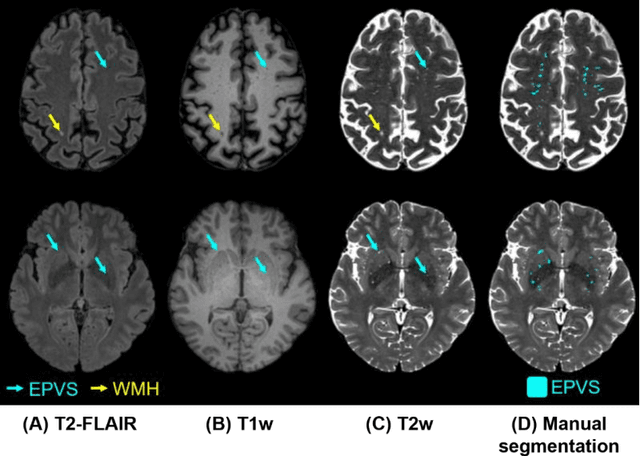
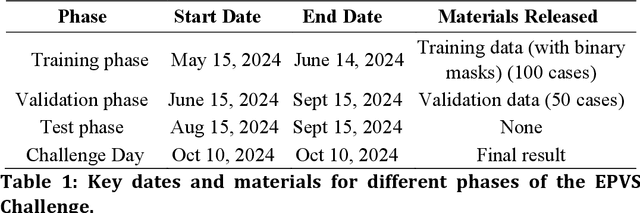
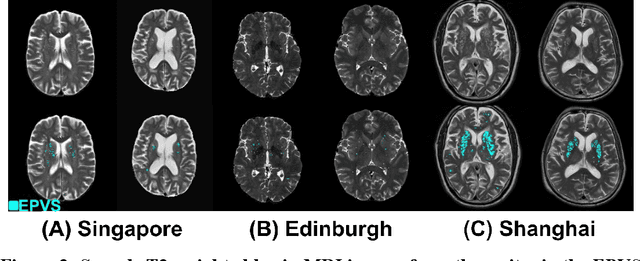
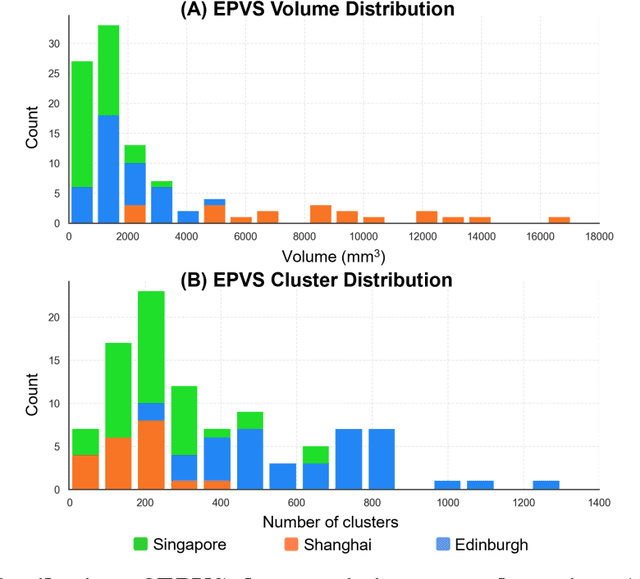
Abstract:Perivascular spaces (PVS), when abnormally enlarged and visible in magnetic resonance imaging (MRI) structural sequences, are important imaging markers of cerebral small vessel disease and potential indicators of neurodegenerative conditions. Despite their clinical significance, automatic enlarged PVS (EPVS) segmentation remains challenging due to their small size, variable morphology, similarity with other pathological features, and limited annotated datasets. This paper presents the EPVS Challenge organized at MICCAI 2024, which aims to advance the development of automated algorithms for EPVS segmentation across multi-site data. We provided a diverse dataset comprising 100 training, 50 validation, and 50 testing scans collected from multiple international sites (UK, Singapore, and China) with varying MRI protocols and demographics. All annotations followed the STRIVE protocol to ensure standardized ground truth and covered the full brain parenchyma. Seven teams completed the full challenge, implementing various deep learning approaches primarily based on U-Net architectures with innovations in multi-modal processing, ensemble strategies, and transformer-based components. Performance was evaluated using dice similarity coefficient, absolute volume difference, recall, and precision metrics. The winning method employed MedNeXt architecture with a dual 2D/3D strategy for handling varying slice thicknesses. The top solutions showed relatively good performance on test data from seen datasets, but significant degradation of performance was observed on the previously unseen Shanghai cohort, highlighting cross-site generalization challenges due to domain shift. This challenge establishes an important benchmark for EPVS segmentation methods and underscores the need for the continued development of robust algorithms that can generalize in diverse clinical settings.
Calibrated Self-supervised Vision Transformers Improve Intracranial Arterial Calcification Segmentation from Clinical CT Head Scans
Jul 02, 2025Abstract:Vision Transformers (ViTs) have gained significant popularity in the natural image domain but have been less successful in 3D medical image segmentation. Nevertheless, 3D ViTs are particularly interesting for large medical imaging volumes due to their efficient self-supervised training within the masked autoencoder (MAE) framework, which enables the use of imaging data without the need for expensive manual annotations. intracranial arterial calcification (IAC) is an imaging biomarker visible on routinely acquired CT scans linked to neurovascular diseases such as stroke and dementia, and automated IAC quantification could enable their large-scale risk assessment. We pre-train ViTs with MAE and fine-tune them for IAC segmentation for the first time. To develop our models, we use highly heterogeneous data from a large clinical trial, the third International Stroke Trial (IST-3). We evaluate key aspects of MAE pre-trained ViTs in IAC segmentation, and analyse the clinical implications. We show: 1) our calibrated self-supervised ViT beats a strong supervised nnU-Net baseline by 3.2 Dice points, 2) low patch sizes are crucial for ViTs for IAC segmentation and interpolation upsampling with regular convolutions is preferable to transposed convolutions for ViT-based models, and 3) our ViTs increase robustness to higher slice thicknesses and improve risk group classification in a clinical scenario by 46%. Our code is available online.
Uncertainty quantification for White Matter Hyperintensity segmentation detects silent failures and improves automated Fazekas quantification
Nov 26, 2024Abstract:White Matter Hyperintensities (WMH) are key neuroradiological markers of small vessel disease present in brain MRI. Assessment of WMH is important in research and clinics. However, WMH are challenging to segment due to their high variability in shape, location, size, poorly defined borders, and similar intensity profile to other pathologies (e.g stroke lesions) and artefacts (e.g head motion). In this work, we apply the most effective techniques for uncertainty quantification (UQ) in segmentation to the WMH segmentation task across multiple test-time data distributions. We find a combination of Stochastic Segmentation Networks with Deep Ensembles yields the highest Dice and lowest Absolute Volume Difference % (AVD) score on in-domain and out-of-distribution data. We demonstrate the downstream utility of UQ, proposing a novel method for classification of the clinical Fazekas score using spatial features extracted for WMH segmentation and UQ maps. We show that incorporating WMH uncertainty information improves Fazekas classification performance and calibration, with median class balanced accuracy for classification models with (UQ and spatial WMH features)/(spatial WMH features)/(WMH volume only) of 0.71/0.66/0.60 in the Deep WMH and 0.82/0.77/0.73 in the Periventricular WMH regions respectively. We demonstrate that stochastic UQ techniques with high sample diversity can improve the detection of poor quality segmentations. Finally, we qualitatively analyse the semantic information captured by UQ techniques and demonstrate that uncertainty can highlight areas where there is ambiguity between WMH and stroke lesions, while identifying clusters of small WMH in deep white matter unsegmented by the model.
Automated neuroradiological support systems for multiple cerebrovascular disease markers -- A systematic review and meta-analysis
Oct 22, 2024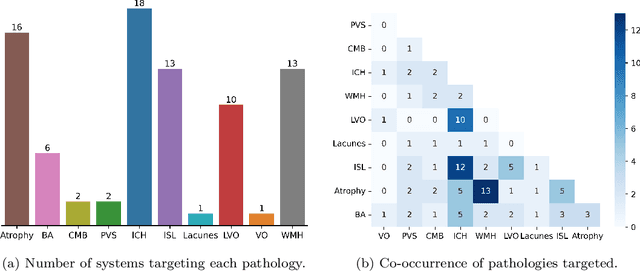
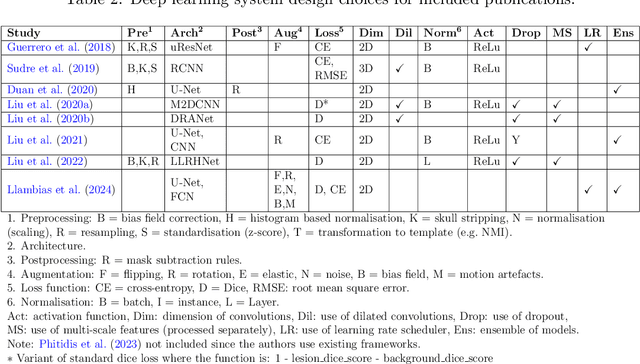
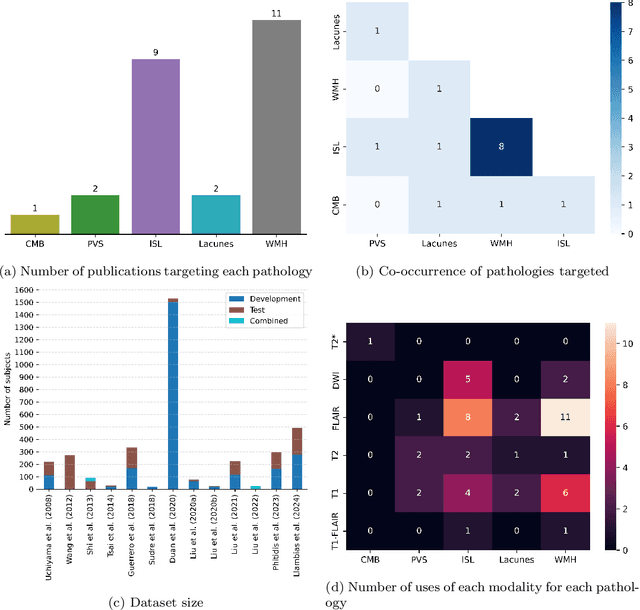
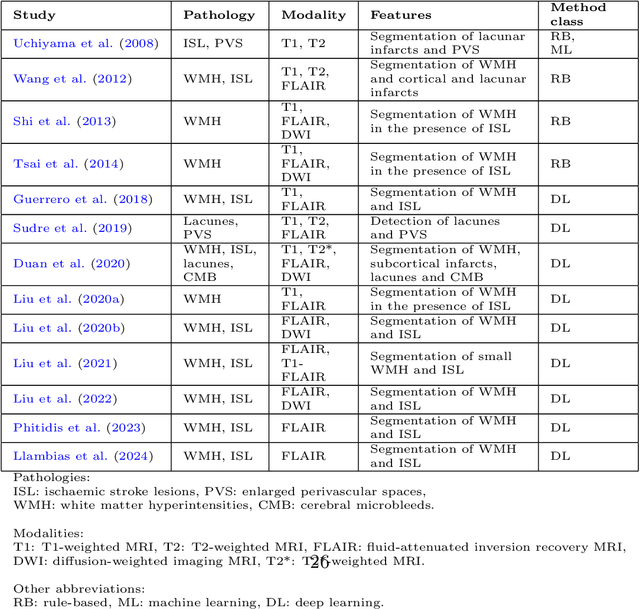
Abstract:Cerebrovascular diseases (CVD) can lead to stroke and dementia. Stroke is the second leading cause of death world wide and dementia incidence is increasing by the year. There are several markers of CVD that are visible on brain imaging, including: white matter hyperintensities (WMH), acute and chronic ischaemic stroke lesions (ISL), lacunes, enlarged perivascular spaces (PVS), acute and chronic haemorrhagic lesions, and cerebral microbleeds (CMB). Brain atrophy also occurs in CVD. These markers are important for patient management and intervention, since they indicate elevated risk of future stroke and dementia. We systematically reviewed automated systems designed to support radiologists reporting on these CVD imaging findings. We considered commercially available software and research publications which identify at least two CVD markers. In total, we included 29 commercial products and 13 research publications. Two distinct types of commercial support system were available: those which identify acute stroke lesions (haemorrhagic and ischaemic) from computed tomography (CT) scans, mainly for the purpose of patient triage; and those which measure WMH and atrophy regionally and longitudinally. In research, WMH and ISL were the markers most frequently analysed together, from magnetic resonance imaging (MRI) scans; lacunes and PVS were each targeted only twice and CMB only once. For stroke, commercially available systems largely support the emergency setting, whilst research systems consider also follow-up and routine scans. The systems to quantify WMH and atrophy are focused on neurodegenerative disease support, where these CVD markers are also of significance. There are currently no openly validated systems, commercially, or in research, performing a comprehensive joint analysis of all CVD markers (WMH, ISL, lacunes, PVS, haemorrhagic lesions, CMB, and atrophy).
Pre-processing and quality control of large clinical CT head datasets for intracranial arterial calcification segmentation
Aug 02, 2024Abstract:As a potential non-invasive biomarker for ischaemic stroke, intracranial arterial calcification (IAC) could be used for stroke risk assessment on CT head scans routinely acquired for other reasons (e.g. trauma, confusion). Artificial intelligence methods can support IAC scoring, but they have not yet been developed for clinical imaging. Large heterogeneous clinical CT datasets are necessary for the training of such methods, but they exhibit expected and unexpected data anomalies. Using CTs from a large clinical trial, the third International Stroke Trial (IST-3), we propose a pipeline that uses as input non-enhanced CT scans to output regions of interest capturing selected large intracranial arteries for IAC scoring. Our method uses co-registration with templates. We focus on quality control, using information presence along the z-axis of the imaging to group and apply similarity measures (structural similarity index measure) to triage assessment of individual image series. Additionally, we propose superimposing thresholded binary masks of the series to inspect large quantities of data in parallel. We identify and exclude unrecoverable samples and registration failures. In total, our pipeline processes 10,659 CT series, rejecting 4,322 (41%) in the entire process, 1,450 (14% of the total) during quality control, and outputting 6,337 series. Our pipeline enables effective and efficient region of interest localisation for targeted IAC segmentation.
Direct Estimation of Pharmacokinetic Parameters from DCE-MRI using Deep CNN with Forward Physical Model Loss
Jun 12, 2018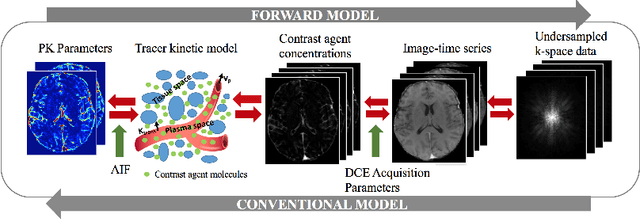
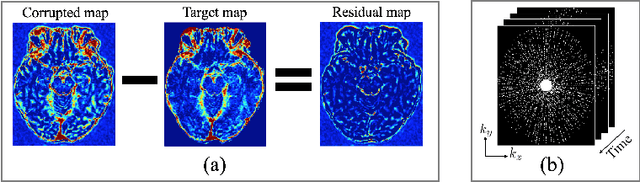
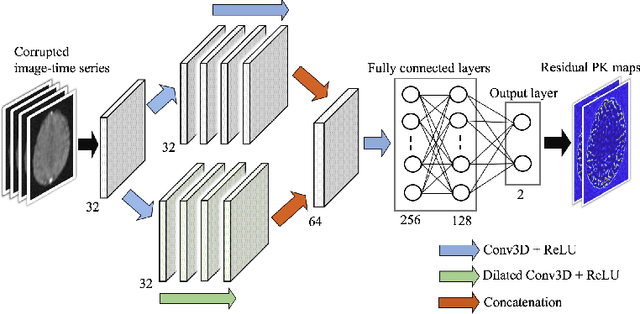
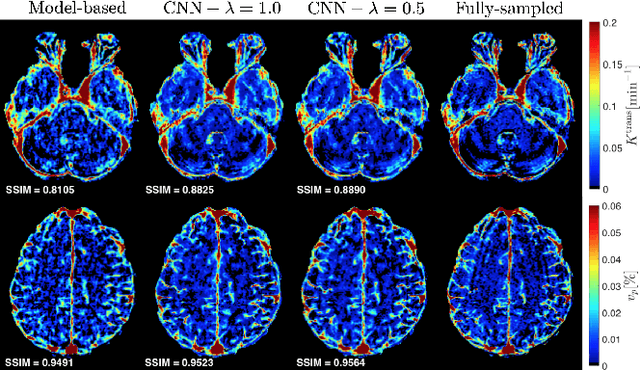
Abstract:Dynamic contrast-enhanced (DCE) MRI is an evolving imaging technique that provides a quantitative measure of pharmacokinetic (PK) parameters in body tissues, in which series of T1-weighted images are collected following the administration of a paramagnetic contrast agent. Unfortunately, in many applications, conventional clinical DCE-MRI suffers from low spatiotemporal resolution and insufficient volume coverage. In this paper, we propose a novel deep learning based approach to directly estimate the PK parameters from undersampled DCE-MRI data. Specifically, we design a custom loss function where we incorporate a forward physical model that relates the PK parameters to corrupted image-time series obtained due to subsampling in k-space. This allows the network to directly exploit the knowledge of true contrast agent kinetics in the training phase, and hence provide more accurate restoration of PK parameters. Experiments on clinical brain DCE datasets demonstrate the efficacy of our approach in terms of fidelity of PK parameter reconstruction and significantly faster parameter inference compared to a model-based iterative reconstruction method.
Perivascular Spaces Segmentation in Brain MRI Using Optimal 3D Filtering
Apr 25, 2017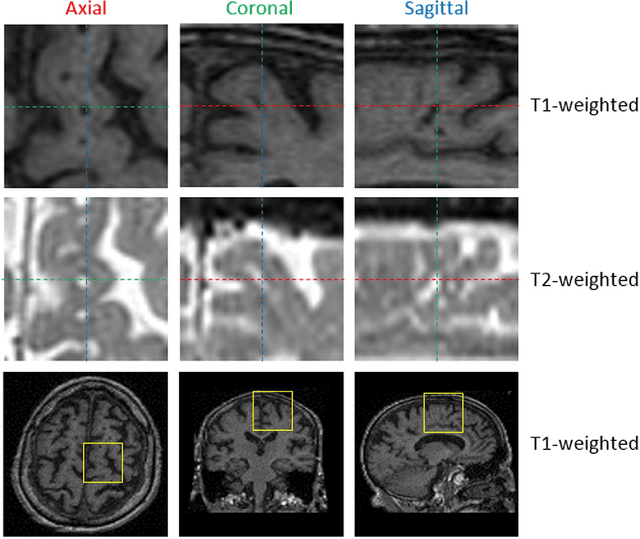
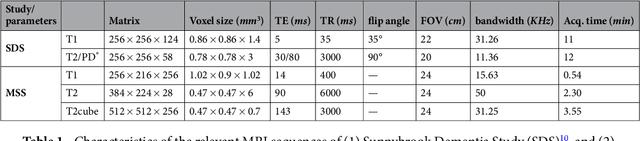
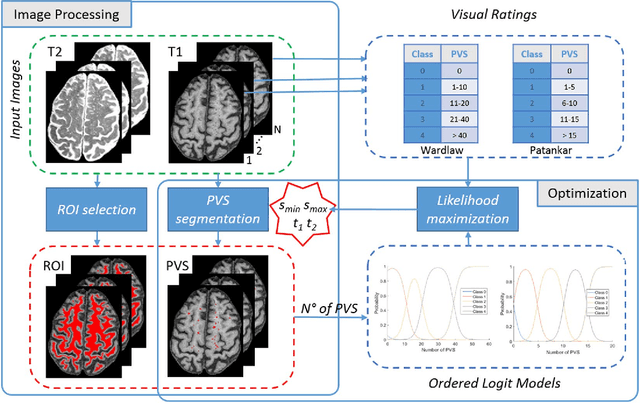
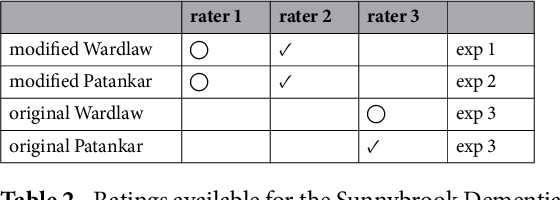
Abstract:Perivascular Spaces (PVS) are a recently recognised feature of Small Vessel Disease (SVD), also indicating neuroinflammation, and are an important part of the brain's circulation and glymphatic drainage system. Quantitative analysis of PVS on Magnetic Resonance Images (MRI) is important for understanding their relationship with neurological diseases. In this work, we propose a segmentation technique based on the 3D Frangi filtering for extraction of PVS from MRI. Based on prior knowledge from neuroradiological ratings of PVS, we used ordered logit models to optimise Frangi filter parameters in response to the variability in the scanner's parameters and study protocols. We optimized and validated our proposed models on two independent cohorts, a dementia sample (N=20) and patients who previously had mild to moderate stroke (N=48). Results demonstrate the robustness and generalisability of our segmentation method. Segmentation-based PVS burden estimates correlated with neuroradiological assessments (Spearman's $\rho$ = 0.74, p $<$ 0.001), suggesting the great potential of our proposed method
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge