Bjoern H. Menze
*: shared first/last authors
Improved Patch Denoising Diffusion Probabilistic Models for Magnetic Resonance Fingerprinting
Oct 29, 2024

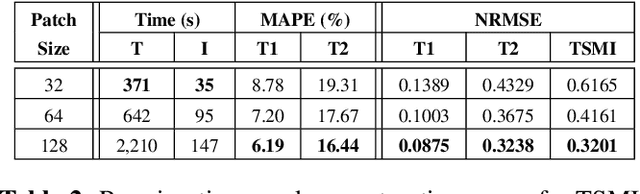

Abstract:Magnetic Resonance Fingerprinting (MRF) is a time-efficient approach to quantitative MRI, enabling the mapping of multiple tissue properties from a single, accelerated scan. However, achieving accurate reconstructions remains challenging, particularly in highly accelerated and undersampled acquisitions, which are crucial for reducing scan times. While deep learning techniques have advanced image reconstruction, the recent introduction of diffusion models offers new possibilities for imaging tasks, though their application in the medical field is still emerging. Notably, diffusion models have not yet been explored for the MRF problem. In this work, we propose for the first time a conditional diffusion probabilistic model for MRF image reconstruction. Qualitative and quantitative comparisons on in-vivo brain scan data demonstrate that the proposed approach can outperform established deep learning and compressed sensing algorithms for MRF reconstruction. Extensive ablation studies also explore strategies to improve computational efficiency of our approach.
StoDIP: Efficient 3D MRF image reconstruction with deep image priors and stochastic iterations
Aug 05, 2024


Abstract:Magnetic Resonance Fingerprinting (MRF) is a time-efficient approach to quantitative MRI for multiparametric tissue mapping. The reconstruction of quantitative maps requires tailored algorithms for removing aliasing artefacts from the compressed sampled MRF acquisitions. Within approaches found in the literature, many focus solely on two-dimensional (2D) image reconstruction, neglecting the extension to volumetric (3D) scans despite their higher relevance and clinical value. A reason for this is that transitioning to 3D imaging without appropriate mitigations presents significant challenges, including increased computational cost and storage requirements, and the need for large amount of ground-truth (artefact-free) data for training. To address these issues, we introduce StoDIP, a new algorithm that extends the ground-truth-free Deep Image Prior (DIP) reconstruction to 3D MRF imaging. StoDIP employs memory-efficient stochastic updates across the multicoil MRF data, a carefully selected neural network architecture, as well as faster nonuniform FFT (NUFFT) transformations. This enables a faster convergence compared against a conventional DIP implementation without these features. Tested on a dataset of whole-brain scans from healthy volunteers, StoDIP demonstrated superior performance over the ground-truth-free reconstruction baselines, both quantitatively and qualitatively.
Deep Image Priors for Magnetic Resonance Fingerprinting with pretrained Bloch-consistent denoising autoencoders
Jul 29, 2024



Abstract:The estimation of multi-parametric quantitative maps from Magnetic Resonance Fingerprinting (MRF) compressed sampled acquisitions, albeit successful, remains a challenge due to the high underspampling rate and artifacts naturally occuring during image reconstruction. Whilst state-of-the-art DL methods can successfully address the task, to fully exploit their capabilities they often require training on a paired dataset, in an area where ground truth is seldom available. In this work, we propose a method that combines a deep image prior (DIP) module that, without ground truth and in conjunction with a Bloch consistency enforcing autoencoder, can tackle the problem, resulting in a method faster and of equivalent or better accuracy than DIP-MRF.
Denoising Diffusion Models for 3D Healthy Brain Tissue Inpainting
Mar 21, 2024



Abstract:Monitoring diseases that affect the brain's structural integrity requires automated analysis of magnetic resonance (MR) images, e.g., for the evaluation of volumetric changes. However, many of the evaluation tools are optimized for analyzing healthy tissue. To enable the evaluation of scans containing pathological tissue, it is therefore required to restore healthy tissue in the pathological areas. In this work, we explore and extend denoising diffusion models for consistent inpainting of healthy 3D brain tissue. We modify state-of-the-art 2D, pseudo-3D, and 3D methods working in the image space, as well as 3D latent and 3D wavelet diffusion models, and train them to synthesize healthy brain tissue. Our evaluation shows that the pseudo-3D model performs best regarding the structural-similarity index, peak signal-to-noise ratio, and mean squared error. To emphasize the clinical relevance, we fine-tune this model on data containing synthetic MS lesions and evaluate it on a downstream brain tissue segmentation task, whereby it outperforms the established FMRIB Software Library (FSL) lesion-filling method.
Framing image registration as a landmark detection problem for better representation of clinical relevance
Jul 31, 2023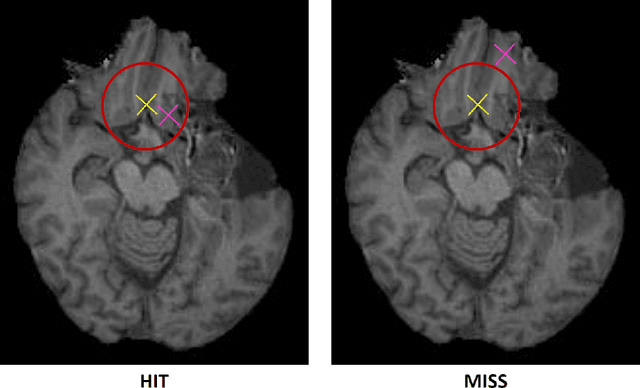
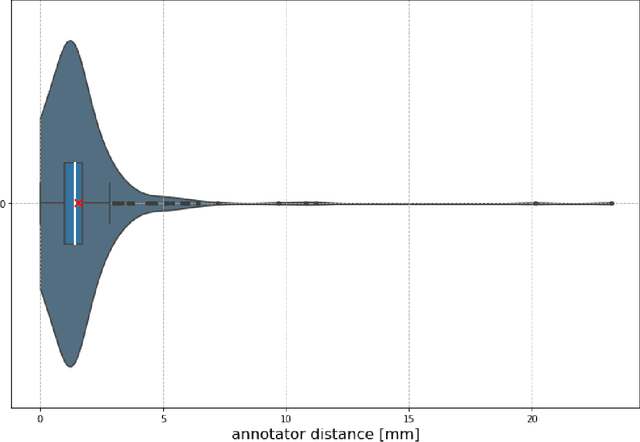
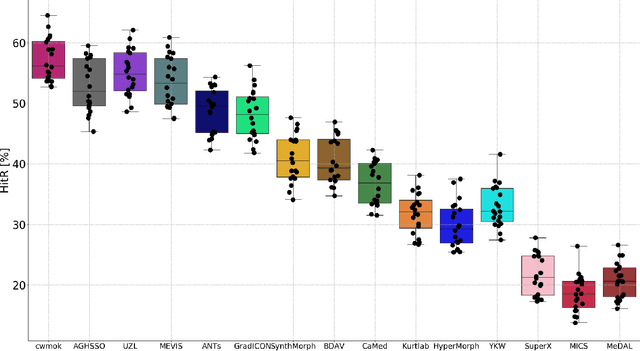
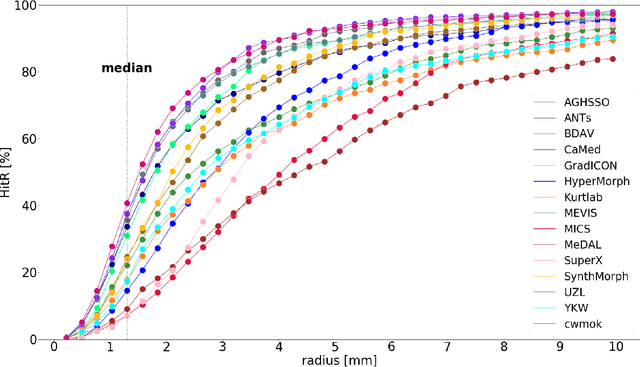
Abstract:Nowadays, registration methods are typically evaluated based on sub-resolution tracking error differences. In an effort to reinfuse this evaluation process with clinical relevance, we propose to reframe image registration as a landmark detection problem. Ideally, landmark-specific detection thresholds are derived from an inter-rater analysis. To approximate this costly process, we propose to compute hit rate curves based on the distribution of errors of a sub-sample inter-rater analysis. Therefore, we suggest deriving thresholds from the error distribution using the formula: median + delta * median absolute deviation. The method promises differentiation of previously indistinguishable registration algorithms and further enables assessing the clinical significance in algorithm development.
Detailed retinal vessel segmentation without human annotations using simulated optical coherence tomography angiographs
Jun 19, 2023Abstract:Optical coherence tomography angiography (OCTA) is a non-invasive imaging modality that can acquire high-resolution volumes of the retinal vasculature and aid the diagnosis of ocular, neurological and cardiac diseases. Segmentation of the visible blood vessels is a common first step when extracting quantitative biomarkers from these images. Classical segmentation algorithms based on thresholding are strongly affected by image artifacts and limited signal-to-noise ratio. The use of modern, deep learning-based segmentation methods has been inhibited by a lack of large datasets with detailed annotations of the blood vessels. To address this issue, recent work has employed transfer learning, where a segmentation network is trained on synthetic OCTA images and is then applied to real data. However, the previously proposed simulation models are incapable of faithfully modeling the retinal vasculature and do not provide effective domain adaptation. Because of this, current methods are not able to fully segment the retinal vasculature, in particular the smallest capillaries. In this work, we present a lightweight simulation of the retinal vascular network based on space colonization for faster and more realistic OCTA synthesis. Moreover, we introduce three contrast adaptation pipelines to decrease the domain gap between real and artificial images. We demonstrate the superior performance of our approach in extensive quantitative and qualitative experiments on three public datasets that compare our method to traditional computer vision algorithms and supervised training using human annotations. Finally, we make our entire pipeline publicly available, including the source code, pretrained models, and a large dataset of synthetic OCTA images.
Primitive Simultaneous Optimization of Similarity Metrics for Image Registration
Apr 04, 2023



Abstract:Even though simultaneous optimization of similarity metrics represents a standard procedure in the field of semantic segmentation, surprisingly, this does not hold true for image registration. To close this unexpected gap in the literature, we investigate in a complex multi-modal 3D setting whether simultaneous optimization of registration metrics, here implemented by means of primitive summation, can benefit image registration. We evaluate two challenging datasets containing collections of pre- to post-operative and pre- to intra-operative Magnetic Resonance Imaging (MRI) of glioma. Employing the proposed optimization we demonstrate improved registration accuracy in terms of Target Registration Error (TRE) on expert neuroradiologists' landmark annotations.
Physiology-based simulation of the retinal vasculature enables annotation-free segmentation of OCT angiographs
Jul 22, 2022



Abstract:Optical coherence tomography angiography (OCTA) can non-invasively image the eye's circulatory system. In order to reliably characterize the retinal vasculature, there is a need to automatically extract quantitative metrics from these images. The calculation of such biomarkers requires a precise semantic segmentation of the blood vessels. However, deep-learning-based methods for segmentation mostly rely on supervised training with voxel-level annotations, which are costly to obtain. In this work, we present a pipeline to synthesize large amounts of realistic OCTA images with intrinsically matching ground truth labels; thereby obviating the need for manual annotation of training data. Our proposed method is based on two novel components: 1) a physiology-based simulation that models the various retinal vascular plexuses and 2) a suite of physics-based image augmentations that emulate the OCTA image acquisition process including typical artifacts. In extensive benchmarking experiments, we demonstrate the utility of our synthetic data by successfully training retinal vessel segmentation algorithms. Encouraged by our method's competitive quantitative and superior qualitative performance, we believe that it constitutes a versatile tool to advance the quantitative analysis of OCTA images.
A unified 3D framework for Organs at Risk Localization and Segmentation for Radiation Therapy Planning
Mar 01, 2022
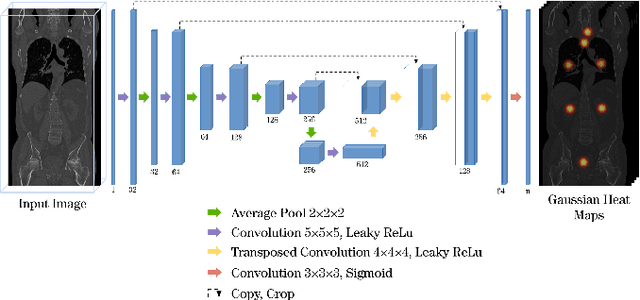


Abstract:Automatic localization and segmentation of organs-at-risk (OAR) in CT are essential pre-processing steps in medical image analysis tasks, such as radiation therapy planning. For instance, the segmentation of OAR surrounding tumors enables the maximization of radiation to the tumor area without compromising the healthy tissues. However, the current medical workflow requires manual delineation of OAR, which is prone to errors and is annotator-dependent. In this work, we aim to introduce a unified 3D pipeline for OAR localization-segmentation rather than novel localization or segmentation architectures. To the best of our knowledge, our proposed framework fully enables the exploitation of 3D context information inherent in medical imaging. In the first step, a 3D multi-variate regression network predicts organs' centroids and bounding boxes. Secondly, 3D organ-specific segmentation networks are leveraged to generate a multi-organ segmentation map. Our method achieved an overall Dice score of $0.9260\pm 0.18 \%$ on the VISCERAL dataset containing CT scans with varying fields of view and multiple organs.
A Deep Learning Approach to Predicting Collateral Flow in Stroke Patients Using Radiomic Features from Perfusion Images
Oct 24, 2021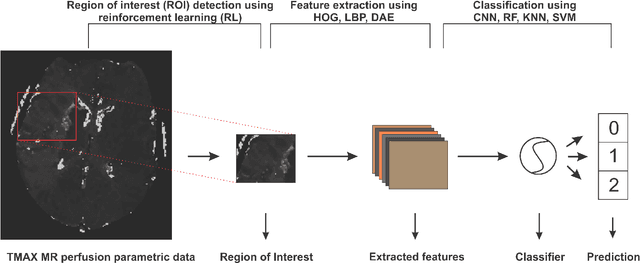
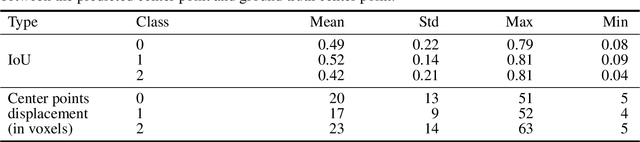
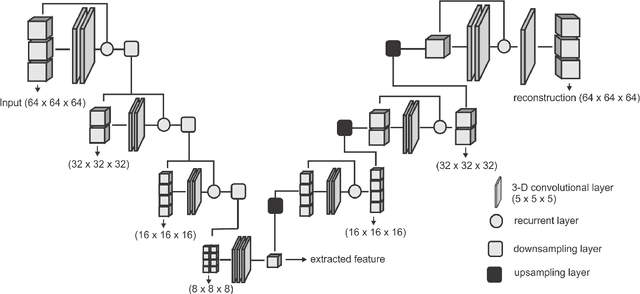
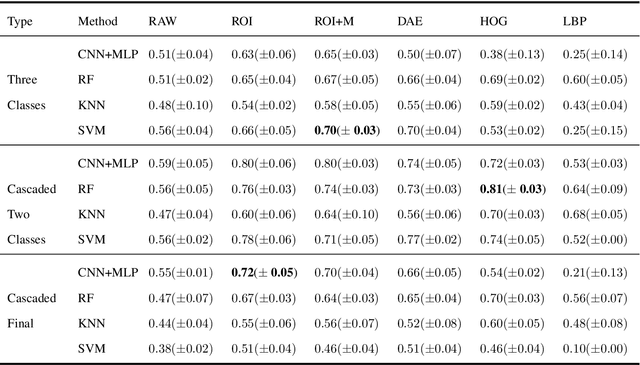
Abstract:Collateral circulation results from specialized anastomotic channels which are capable of providing oxygenated blood to regions with compromised blood flow caused by ischemic injuries. The quality of collateral circulation has been established as a key factor in determining the likelihood of a favorable clinical outcome and goes a long way to determine the choice of stroke care model - that is the decision to transport or treat eligible patients immediately. Though there exist several imaging methods and grading criteria for quantifying collateral blood flow, the actual grading is mostly done through manual inspection of the acquired images. This approach is associated with a number of challenges. First, it is time-consuming - the clinician needs to scan through several slices of images to ascertain the region of interest before deciding on what severity grade to assign to a patient. Second, there is a high tendency for bias and inconsistency in the final grade assigned to a patient depending on the experience level of the clinician. We present a deep learning approach to predicting collateral flow grading in stroke patients based on radiomic features extracted from MR perfusion data. First, we formulate a region of interest detection task as a reinforcement learning problem and train a deep learning network to automatically detect the occluded region within the 3D MR perfusion volumes. Second, we extract radiomic features from the obtained region of interest through local image descriptors and denoising auto-encoders. Finally, we apply a convolutional neural network and other machine learning classifiers to the extracted radiomic features to automatically predict the collateral flow grading of the given patient volume as one of three severity classes - no flow (0), moderate flow (1), and good flow (2)...
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge