Marion I. Menzel
on behalf of the PREDICTOM consortium
INR meets Multi-Contrast MRI Reconstruction
Sep 05, 2025Abstract:Multi-contrast MRI sequences allow for the acquisition of images with varying tissue contrast within a single scan. The resulting multi-contrast images can be used to extract quantitative information on tissue microstructure. To make such multi-contrast sequences feasible for clinical routine, the usually very long scan times need to be shortened e.g. through undersampling in k-space. However, this comes with challenges for the reconstruction. In general, advanced reconstruction techniques such as compressed sensing or deep learning-based approaches can enable the acquisition of high-quality images despite the acceleration. In this work, we leverage redundant anatomical information of multi-contrast sequences to achieve even higher acceleration rates. We use undersampling patterns that capture the contrast information located at the k-space center, while performing complementary undersampling across contrasts for high frequencies. To reconstruct this highly sparse k-space data, we propose an implicit neural representation (INR) network that is ideal for using the complementary information acquired across contrasts as it jointly reconstructs all contrast images. We demonstrate the benefits of our proposed INR method by applying it to multi-contrast MRI using the MPnRAGE sequence, where it outperforms the state-of-the-art parallel imaging compressed sensing (PICS) reconstruction method, even at higher acceleration factors.
StoDIP: Efficient 3D MRF image reconstruction with deep image priors and stochastic iterations
Aug 05, 2024


Abstract:Magnetic Resonance Fingerprinting (MRF) is a time-efficient approach to quantitative MRI for multiparametric tissue mapping. The reconstruction of quantitative maps requires tailored algorithms for removing aliasing artefacts from the compressed sampled MRF acquisitions. Within approaches found in the literature, many focus solely on two-dimensional (2D) image reconstruction, neglecting the extension to volumetric (3D) scans despite their higher relevance and clinical value. A reason for this is that transitioning to 3D imaging without appropriate mitigations presents significant challenges, including increased computational cost and storage requirements, and the need for large amount of ground-truth (artefact-free) data for training. To address these issues, we introduce StoDIP, a new algorithm that extends the ground-truth-free Deep Image Prior (DIP) reconstruction to 3D MRF imaging. StoDIP employs memory-efficient stochastic updates across the multicoil MRF data, a carefully selected neural network architecture, as well as faster nonuniform FFT (NUFFT) transformations. This enables a faster convergence compared against a conventional DIP implementation without these features. Tested on a dataset of whole-brain scans from healthy volunteers, StoDIP demonstrated superior performance over the ground-truth-free reconstruction baselines, both quantitatively and qualitatively.
Deep Image Priors for Magnetic Resonance Fingerprinting with pretrained Bloch-consistent denoising autoencoders
Jul 29, 2024



Abstract:The estimation of multi-parametric quantitative maps from Magnetic Resonance Fingerprinting (MRF) compressed sampled acquisitions, albeit successful, remains a challenge due to the high underspampling rate and artifacts naturally occuring during image reconstruction. Whilst state-of-the-art DL methods can successfully address the task, to fully exploit their capabilities they often require training on a paired dataset, in an area where ground truth is seldom available. In this work, we propose a method that combines a deep image prior (DIP) module that, without ground truth and in conjunction with a Bloch consistency enforcing autoencoder, can tackle the problem, resulting in a method faster and of equivalent or better accuracy than DIP-MRF.
With or Without Replacement? Improving Confidence in Fourier Imaging
Jul 18, 2024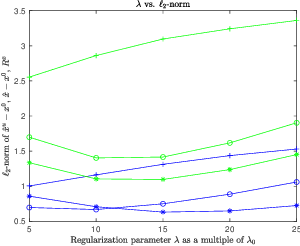
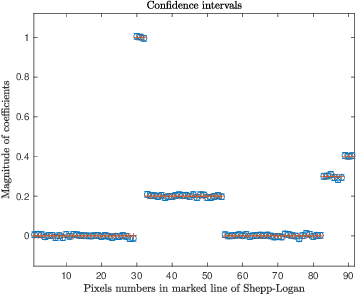
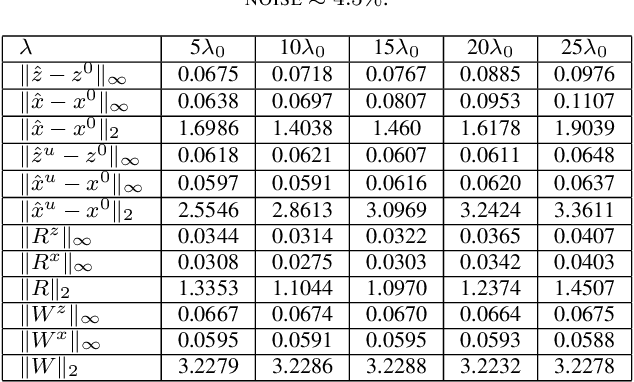
Abstract:Over the last few years, debiased estimators have been proposed in order to establish rigorous confidence intervals for high-dimensional problems in machine learning and data science. The core argument is that the error of these estimators with respect to the ground truth can be expressed as a Gaussian variable plus a remainder term that vanishes as long as the dimension of the problem is sufficiently high. Thus, uncertainty quantification (UQ) can be performed exploiting the Gaussian model. Empirically, however, the remainder term cannot be neglected in many realistic situations of moderately-sized dimensions, in particular in certain structured measurement scenarios such as Magnetic Resonance Imaging (MRI). This, in turn, can downgrade the advantage of the UQ methods as compared to non-UQ approaches such as the standard LASSO. In this paper, we present a method to improve the debiased estimator by sampling without replacement. Our approach leverages recent results of ours on the structure of the random nature of certain sampling schemes showing how a transition between sampling with and without replacement can lead to a weighted reconstruction scheme with improved performance for the standard LASSO. In this paper, we illustrate how this reweighted sampling idea can also improve the debiased estimator and, consequently, provide a better method for UQ in Fourier imaging.
Nonlinear Equivariant Imaging: Learning Multi-Parametric Tissue Mapping without Ground Truth for Compressive Quantitative MRI
Nov 23, 2022Abstract:Current state-of-the-art reconstruction for quantitative tissue maps from fast, compressive, Magnetic Resonance Fingerprinting (MRF), use supervised deep learning, with the drawback of requiring high-fidelity ground truth tissue map training data which is limited. This paper proposes NonLinear Equivariant Imaging (NLEI), a self-supervised learning approach to eliminate the need for ground truth for deep MRF image reconstruction. NLEI extends the recent Equivariant Imaging framework to nonlinear inverse problems such as MRF. Only fast, compressed-sampled MRF scans are used for training. NLEI learns tissue mapping using spatiotemporal priors: spatial priors are obtained from the invariance of MRF data to a group of geometric image transformations, while temporal priors are obtained from a nonlinear Bloch response model approximated by a pre-trained neural network. Tested retrospectively on two acquisition settings, we observe that NLEI (self-supervised learning) closely approaches the performance of supervised learning, despite not using ground truth during training.
A Plug-and-Play Approach to Multiparametric Quantitative MRI: Image Reconstruction using Pre-Trained Deep Denoisers
Feb 10, 2022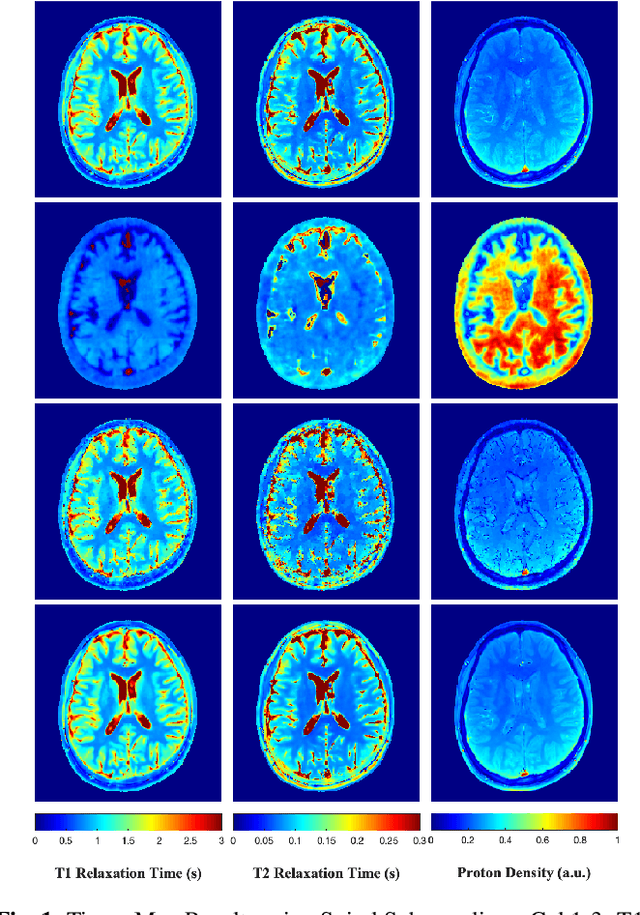
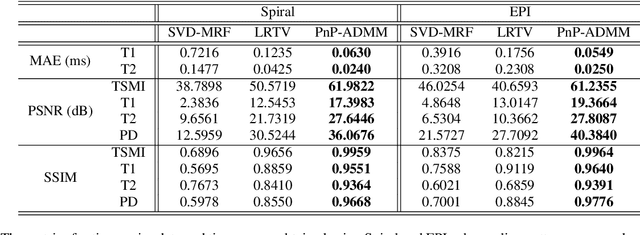
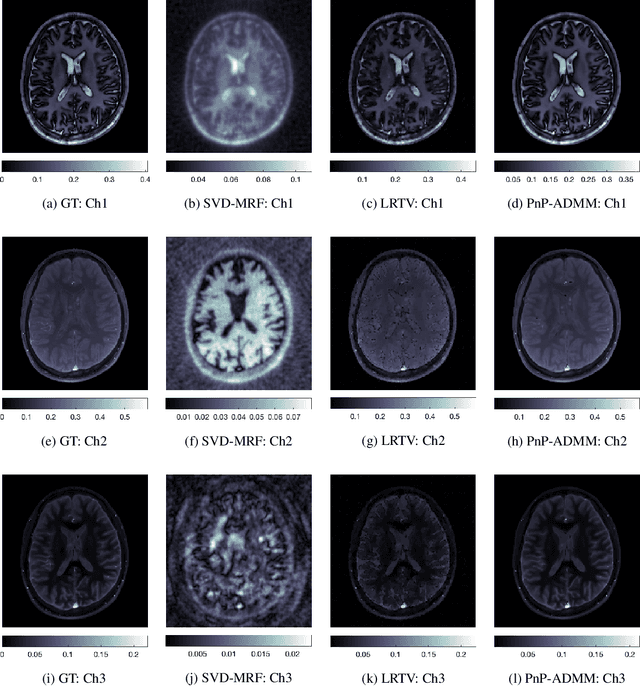
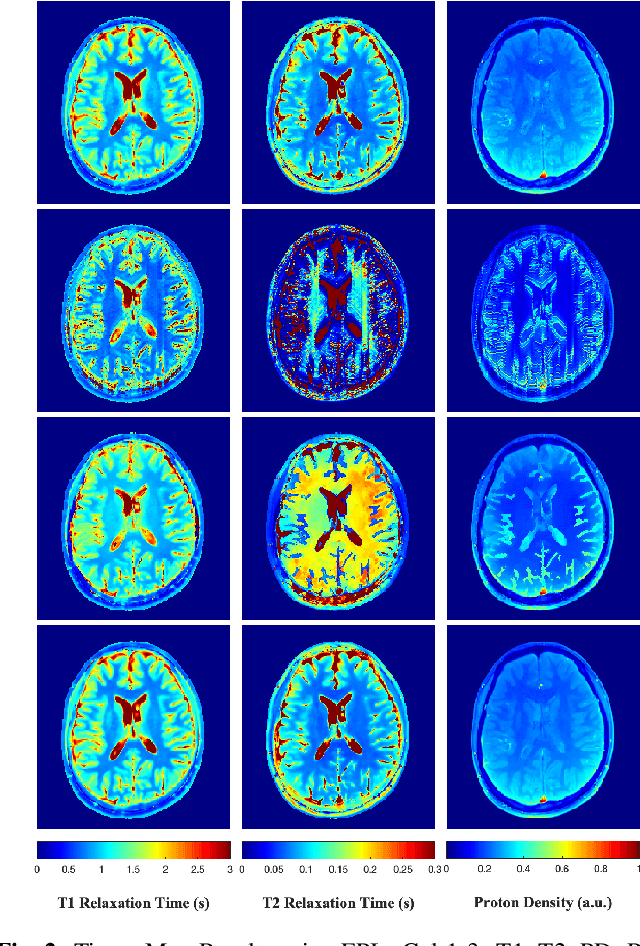
Abstract:Current spatiotemporal deep learning approaches to Magnetic Resonance Fingerprinting (MRF) build artefact-removal models customised to a particular k-space subsampling pattern which is used for fast (compressed) acquisition. This may not be useful when the acquisition process is unknown during training of the deep learning model and/or changes during testing time. This paper proposes an iterative deep learning plug-and-play reconstruction approach to MRF which is adaptive to the forward acquisition process. Spatiotemporal image priors are learned by an image denoiser i.e. a Convolutional Neural Network (CNN), trained to remove generic white gaussian noise (not a particular subsampling artefact) from data. This CNN denoiser is then used as a data-driven shrinkage operator within the iterative reconstruction algorithm. This algorithm with the same denoiser model is then tested on two simulated acquisition processes with distinct subsampling patterns. The results show consistent de-aliasing performance against both acquisition schemes and accurate mapping of tissues' quantitative bio-properties. Software available: https://github.com/ketanfatania/QMRI-PnP-Recon-POC
Data Harmonisation for Information Fusion in Digital Healthcare: A State-of-the-Art Systematic Review, Meta-Analysis and Future Research Directions
Jan 17, 2022

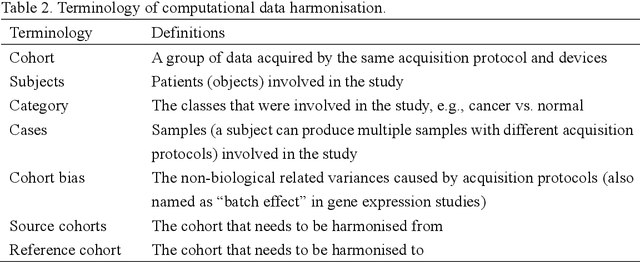

Abstract:Removing the bias and variance of multicentre data has always been a challenge in large scale digital healthcare studies, which requires the ability to integrate clinical features extracted from data acquired by different scanners and protocols to improve stability and robustness. Previous studies have described various computational approaches to fuse single modality multicentre datasets. However, these surveys rarely focused on evaluation metrics and lacked a checklist for computational data harmonisation studies. In this systematic review, we summarise the computational data harmonisation approaches for multi-modality data in the digital healthcare field, including harmonisation strategies and evaluation metrics based on different theories. In addition, a comprehensive checklist that summarises common practices for data harmonisation studies is proposed to guide researchers to report their research findings more effectively. Last but not least, flowcharts presenting possible ways for methodology and metric selection are proposed and the limitations of different methods have been surveyed for future research.
Deep MR Fingerprinting with total-variation and low-rank subspace priors
Feb 26, 2019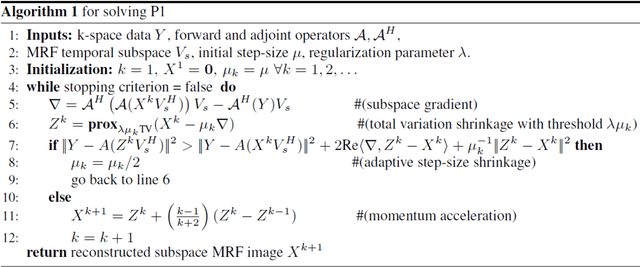
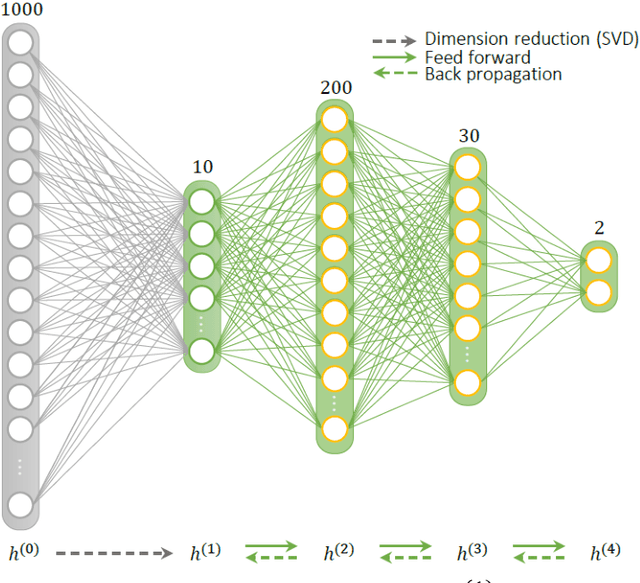
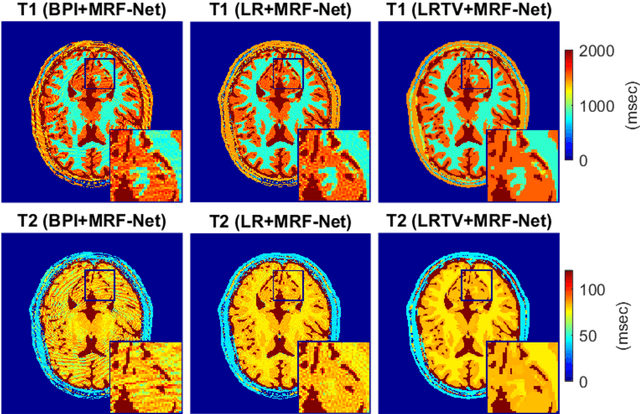
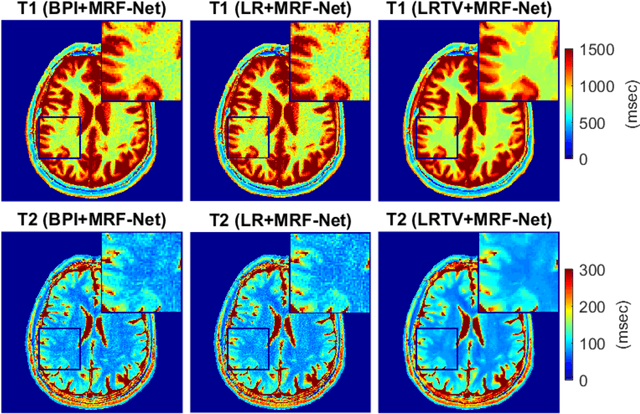
Abstract:Deep learning (DL) has recently emerged to address the heavy storage and computation requirements of the baseline dictionary-matching (DM) for Magnetic Resonance Fingerprinting (MRF) reconstruction. Fed with non-iterated back-projected images, the network is unable to fully resolve spatially-correlated corruptions caused from the undersampling artefacts. We propose an accelerated iterative reconstruction to minimize these artefacts before feeding into the network. This is done through a convex regularization that jointly promotes spatio-temporal regularities of the MRF time-series. Except for training, the rest of the parameter estimation pipeline is dictionary-free. We validate the proposed approach on synthetic and in-vivo datasets.
Geometry of Deep Learning for Magnetic Resonance Fingerprinting
Nov 04, 2018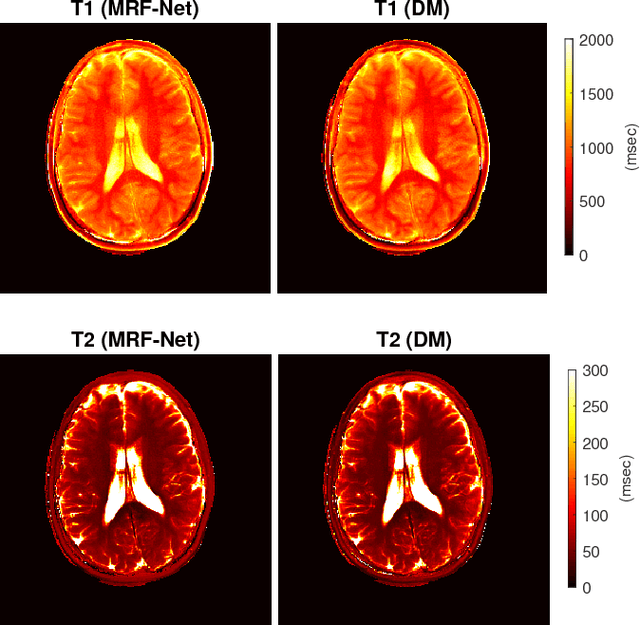
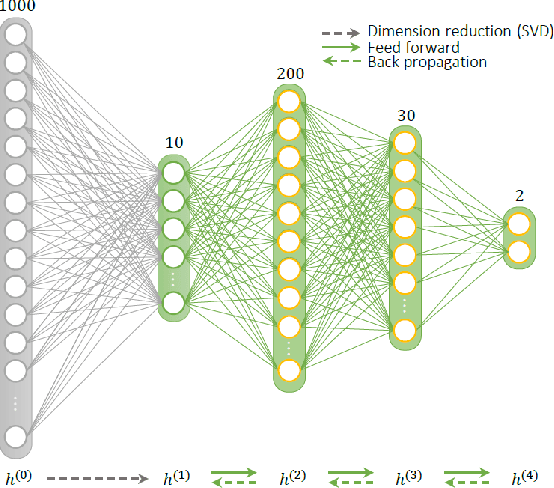
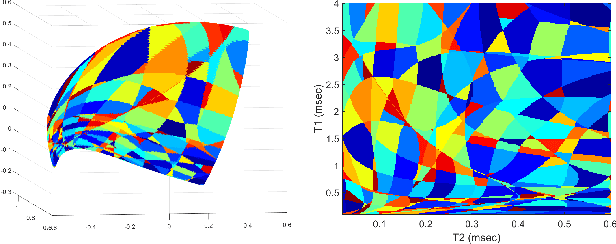
Abstract:Current popular methods for Magnetic Resonance Fingerprint (MRF) recovery are bottlenecked by the heavy storage and computation requirements of a dictionary-matching (DM) step due to the growing size and complexity of the fingerprint dictionaries in multi-parametric quantitative MRI applications. In this paper we study a deep learning approach to address these shortcomings. Coupled with a dimensionality reduction first layer, the proposed MRF-Net is able to reconstruct quantitative maps by saving more than 60 times in memory and computations required for a DM baseline. Fine-grid manifold enumeration i.e. the MRF dictionary is only used for training the network and not during image reconstruction. We show that the MRF-Net provides a piece-wise affine approximation to the Bloch response manifold projection and that rather than memorizing the dictionary, the network efficiently clusters this manifold and learns a set of hierarchical matched-filters for affine regression of the NMR characteristics in each segment.
Balanced multi-shot EPI for accelerated Cartesian MRF: An alternative to spiral MRF
Sep 06, 2018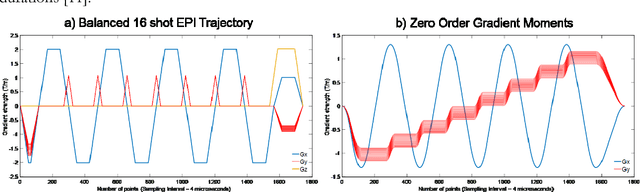
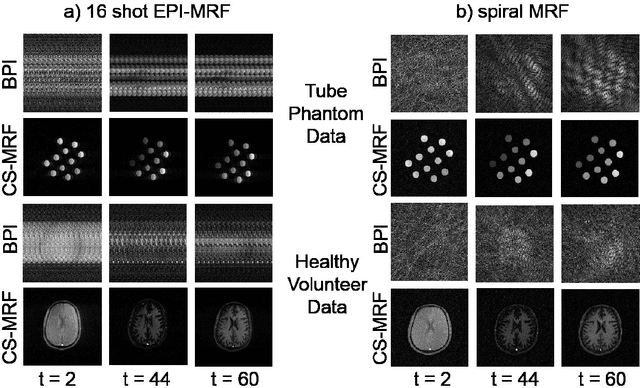
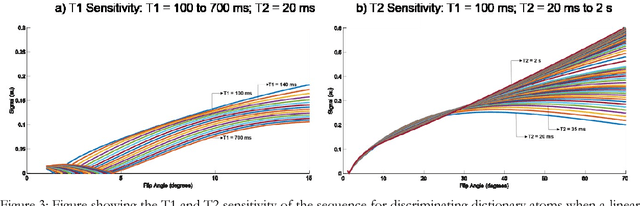
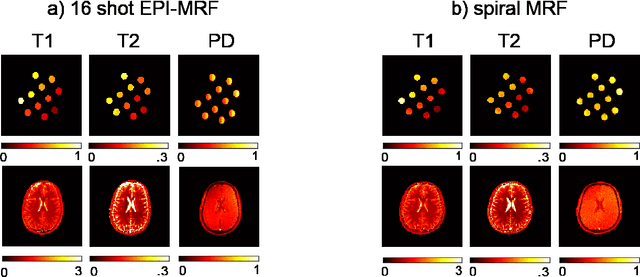
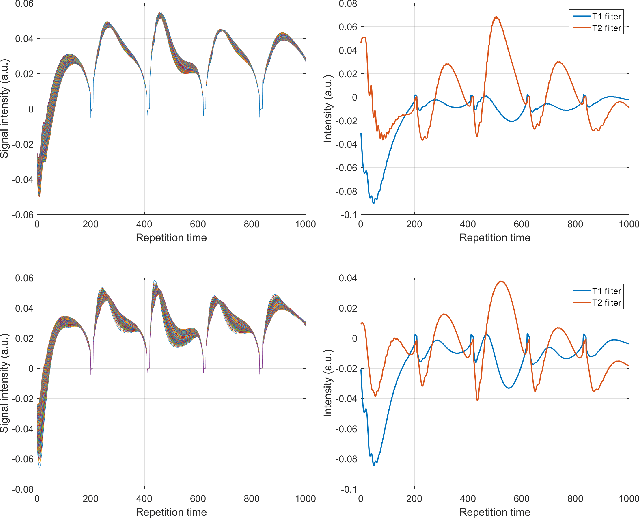
Abstract:The main purpose of this study is to show that a highly accelerated Cartesian MRF scheme using a multi-shot EPI readout (i.e. multi-shot EPI-MRF) can produce good quality multi-parametric maps such as T1, T2 and proton density (PD) in a sufficiently short scan duration that is similar to conventional MRF. This multi-shot approach allows considerable subsampling while traversing the entire k-space trajectory, can yield better SNR, reduced blurring, less distortion and can also be used to collect higher resolution data compared to existing single-shot EPI-MRF implementations. The generated parametric maps are compared to an accelerated spiral MRF implementation with the same acquisition parameters to evaluate the performance of this method. Additionally, an iterative reconstruction algorithm is applied to improve the accuracy of parametric map estimations and the fast convergence of EPI-MRF is also demonstrated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge