Michael Roberts
on behalf of the AIX-COVNET collaboration
Laplace Learning in Wasserstein Space
Nov 17, 2025Abstract:The manifold hypothesis posits that high-dimensional data typically resides on low-dimensional sub spaces. In this paper, we assume manifold hypothesis to investigate graph-based semi-supervised learning methods. In particular, we examine Laplace Learning in the Wasserstein space, extending the classical notion of graph-based semi-supervised learning algorithms from finite-dimensional Euclidean spaces to an infinite-dimensional setting. To achieve this, we prove variational convergence of a discrete graph p- Dirichlet energy to its continuum counterpart. In addition, we characterize the Laplace-Beltrami operator on asubmanifold of the Wasserstein space. Finally, we validate the proposed theoretical framework through numerical experiments conducted on benchmark datasets, demonstrating the consistency of our classification performance in high-dimensional settings.
SurvSurf: a partially monotonic neural network for first-hitting time prediction of intermittently observed discrete and continuous sequential events
Apr 07, 2025Abstract:We propose a neural-network based survival model (SurvSurf) specifically designed for direct and simultaneous probabilistic prediction of the first hitting time of sequential events from baseline. Unlike existing models, SurvSurf is theoretically guaranteed to never violate the monotonic relationship between the cumulative incidence functions of sequential events, while allowing nonlinear influence from predictors. It also incorporates implicit truths for unobserved intermediate events in model fitting, and supports both discrete and continuous time and events. We also identified a variant of the Integrated Brier Score (IBS) that showed robust correlation with the mean squared error (MSE) between the true and predicted probabilities by accounting for implied truths about the missing intermediate events. We demonstrated the superiority of SurvSurf compared to modern and traditional predictive survival models in two simulated datasets and two real-world datasets, using MSE, the more robust IBS and by measuring the extent of monotonicity violation.
Parameter choices in HaarPSI for IQA with medical images
Oct 31, 2024Abstract:When developing machine learning models, image quality assessment (IQA) measures are a crucial component for evaluation. However, commonly used IQA measures have been primarily developed and optimized for natural images. In many specialized settings, such as medical images, this poses an often-overlooked problem regarding suitability. In previous studies, the IQA measure HaarPSI showed promising behavior for natural and medical images. HaarPSI is based on Haar wavelet representations and the framework allows optimization of two parameters. So far, these parameters have been aligned for natural images. Here, we optimize these parameters for two annotated medical data sets, a photoacoustic and a chest X-Ray data set. We observe that they are more sensitive to the parameter choices than the employed natural images, and on the other hand both medical data sets lead to similar parameter values when optimized. We denote the optimized setting, which improves the performance for the medical images notably, by HaarPSI$_{MED}$. The results suggest that adapting common IQA measures within their frameworks for medical images can provide a valuable, generalizable addition to the employment of more specific task-based measures.
Deep Generative Classification of Blood Cell Morphology
Aug 16, 2024



Abstract:Accurate classification of haematological cells is critical for diagnosing blood disorders, but presents significant challenges for machine automation owing to the complexity of cell morphology, heterogeneities of biological, pathological, and imaging characteristics, and the imbalance of cell type frequencies. We introduce CytoDiffusion, a diffusion-based classifier that effectively models blood cell morphology, combining accurate classification with robust anomaly detection, resistance to distributional shifts, interpretability, data efficiency, and superhuman uncertainty quantification. Our approach outperforms state-of-the-art discriminative models in anomaly detection (AUC 0.976 vs. 0.919), resistance to domain shifts (85.85% vs. 74.38% balanced accuracy), and performance in low-data regimes (95.88% vs. 94.95% balanced accuracy). Notably, our model generates synthetic blood cell images that are nearly indistinguishable from real images, as demonstrated by a Turing test in which expert haematologists achieved only 52.3% accuracy (95% CI: [50.5%, 54.2%]). Furthermore, we enhance model explainability through the generation of directly interpretable counterfactual heatmaps. Our comprehensive evaluation framework, encompassing these multiple performance dimensions, establishes a new benchmark for medical image analysis in haematology, ultimately enabling improved diagnostic accuracy in clinical settings. Our code is available at https://github.com/Deltadahl/CytoDiffusion.
FedMAP: Unlocking Potential in Personalized Federated Learning through Bi-Level MAP Optimization
May 29, 2024



Abstract:Federated Learning (FL) enables collaborative training of machine learning models on decentralized data while preserving data privacy. However, data across clients often differs significantly due to class imbalance, feature distribution skew, sample size imbalance, and other phenomena. Leveraging information from these not identically distributed (non-IID) datasets poses substantial challenges. FL methods based on a single global model cannot effectively capture the variations in client data and underperform in non-IID settings. Consequently, Personalized FL (PFL) approaches that adapt to each client's data distribution but leverage other clients' data are essential but currently underexplored. We propose a novel Bayesian PFL framework using bi-level optimization to tackle the data heterogeneity challenges. Our proposed framework utilizes the global model as a prior distribution within a Maximum A Posteriori (MAP) estimation of personalized client models. This approach facilitates PFL by integrating shared knowledge from the prior, thereby enhancing local model performance, generalization ability, and communication efficiency. We extensively evaluated our bi-level optimization approach on real-world and synthetic datasets, demonstrating significant improvements in model accuracy compared to existing methods while reducing communication overhead. This study contributes to PFL by establishing a solid theoretical foundation for the proposed method and offering a robust, ready-to-use framework that effectively addresses the challenges posed by non-IID data in FL.
A study of why we need to reassess full reference image quality assessment with medical images
May 29, 2024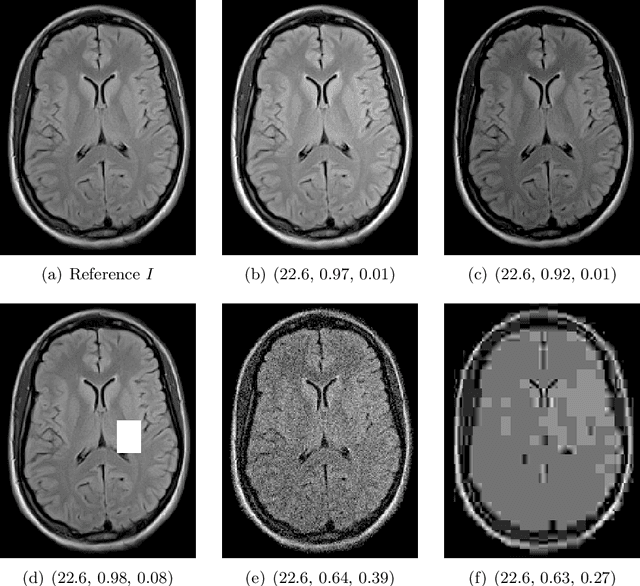
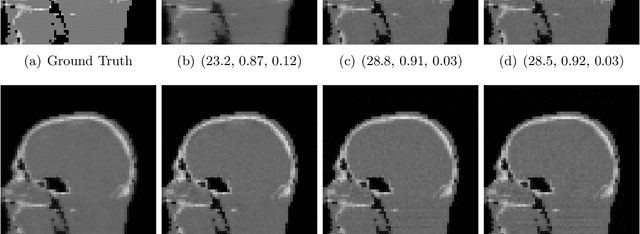
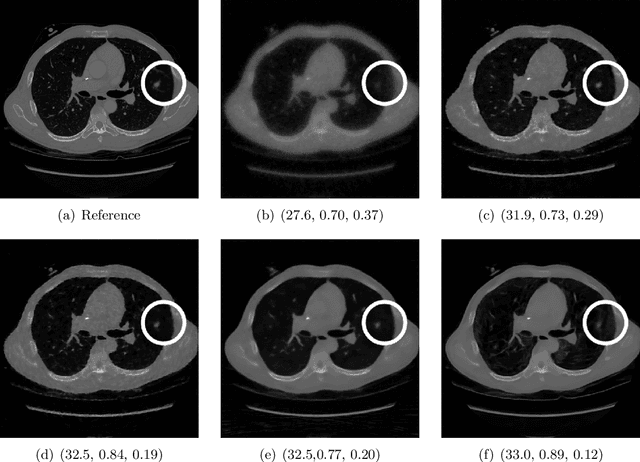
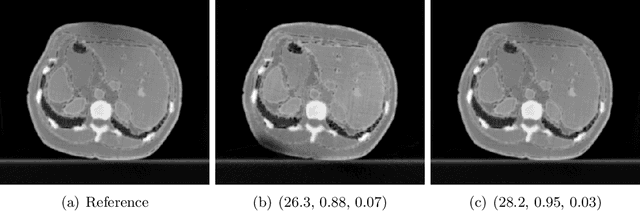
Abstract:Image quality assessment (IQA) is not just indispensable in clinical practice to ensure high standards, but also in the development stage of novel algorithms that operate on medical images with reference data. This paper provides a structured and comprehensive collection of examples where the two most common full reference (FR) image quality measures prove to be unsuitable for the assessment of novel algorithms using different kinds of medical images, including real-world MRI, CT, OCT, X-Ray, digital pathology and photoacoustic imaging data. In particular, the FR-IQA measures PSNR and SSIM are known and tested for working successfully in many natural imaging tasks, but discrepancies in medical scenarios have been noted in the literature. Inconsistencies arising in medical images are not surprising, as they have very different properties than natural images which have not been targeted nor tested in the development of the mentioned measures, and therefore might imply wrong judgement of novel methods for medical images. Therefore, improvement is urgently needed in particular in this era of AI to increase explainability, reproducibility and generalizability in machine learning for medical imaging and beyond. On top of the pitfalls we will provide ideas for future research as well as suggesting guidelines for the usage of FR-IQA measures applied to medical images.
A study on the adequacy of common IQA measures for medical images
May 29, 2024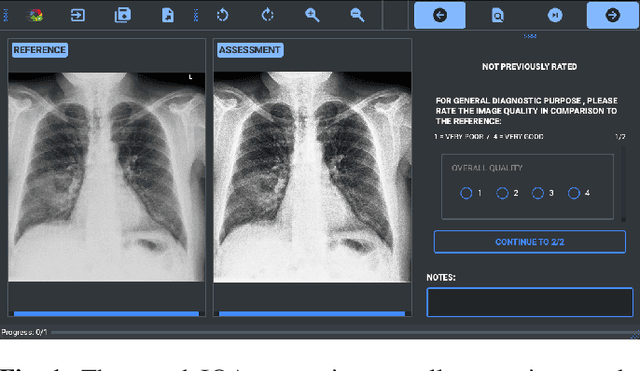
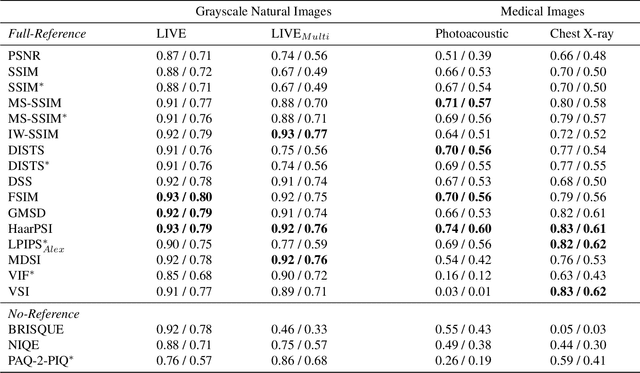
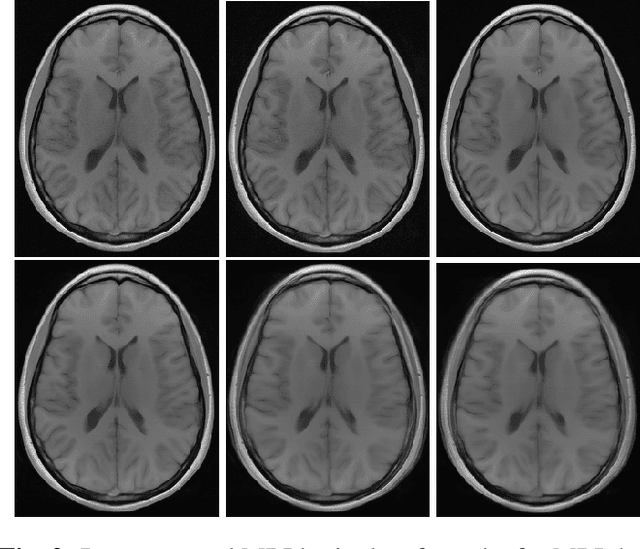
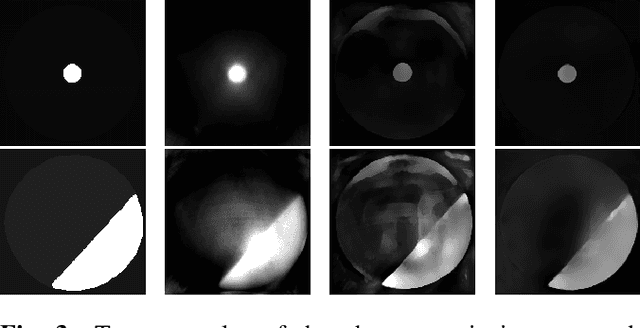
Abstract:Image quality assessment (IQA) is standard practice in the development stage of novel machine learning algorithms that operate on images. The most commonly used IQA measures have been developed and tested for natural images, but not in the medical setting. Reported inconsistencies arising in medical images are not surprising, as they have different properties than natural images. In this study, we test the applicability of common IQA measures for medical image data by comparing their assessment to manually rated chest X-ray (5 experts) and photoacoustic image data (1 expert). Moreover, we include supplementary studies on grayscale natural images and accelerated brain MRI data. The results of all experiments show a similar outcome in line with previous findings for medical imaging: PSNR and SSIM in the default setting are in the lower range of the result list and HaarPSI outperforms the other tested measures in the overall performance. Also among the top performers in our medical experiments are the full reference measures DISTS, FSIM, LPIPS and MS-SSIM. Generally, the results on natural images yield considerably higher correlations, suggesting that the additional employment of tailored IQA measures for medical imaging algorithms is needed.
The curious case of the test set AUROC
Dec 19, 2023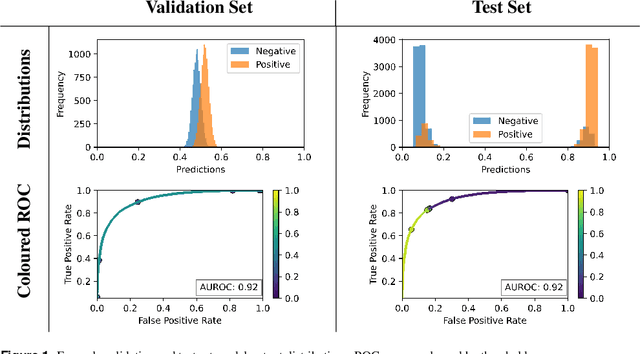
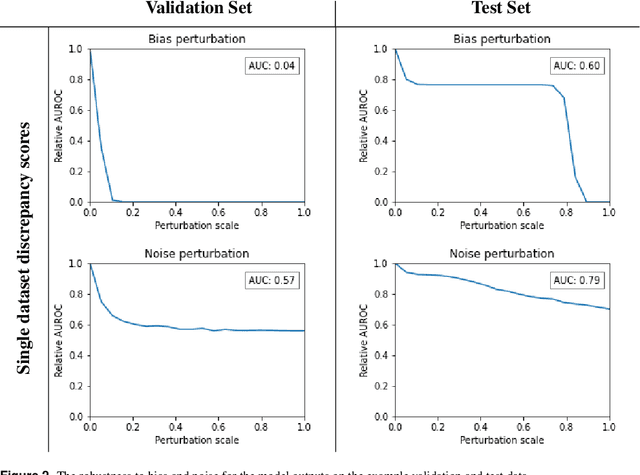
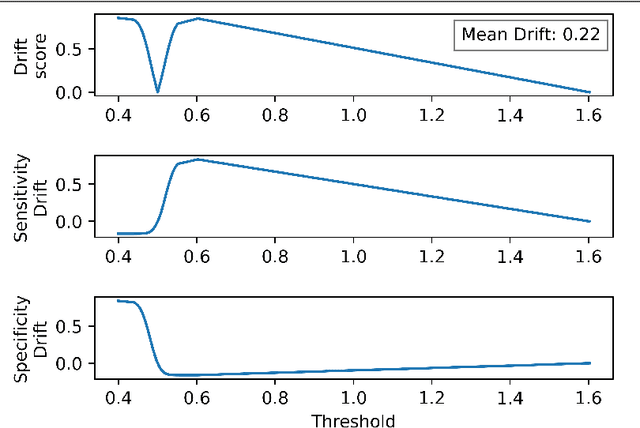
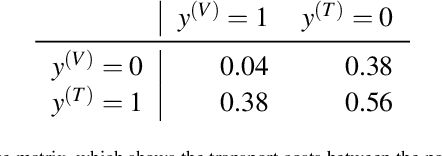
Abstract:Whilst the size and complexity of ML models have rapidly and significantly increased over the past decade, the methods for assessing their performance have not kept pace. In particular, among the many potential performance metrics, the ML community stubbornly continues to use (a) the area under the receiver operating characteristic curve (AUROC) for a validation and test cohort (distinct from training data) or (b) the sensitivity and specificity for the test data at an optimal threshold determined from the validation ROC. However, we argue that considering scores derived from the test ROC curve alone gives only a narrow insight into how a model performs and its ability to generalise.
Recent Methodological Advances in Federated Learning for Healthcare
Oct 04, 2023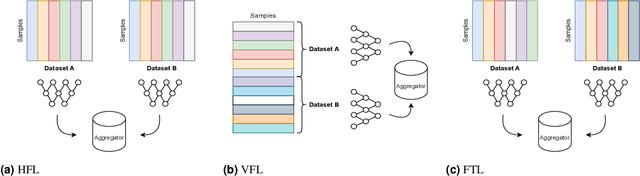
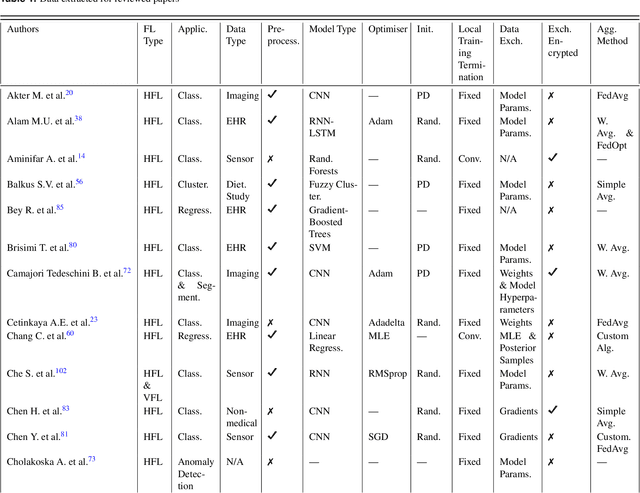
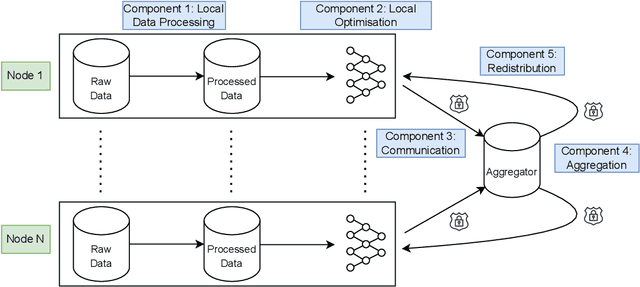
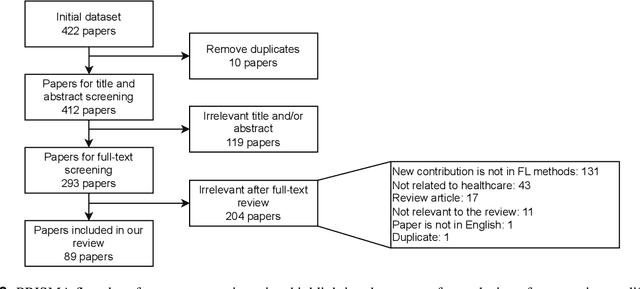
Abstract:For healthcare datasets, it is often not possible to combine data samples from multiple sites due to ethical, privacy or logistical concerns. Federated learning allows for the utilisation of powerful machine learning algorithms without requiring the pooling of data. Healthcare data has many simultaneous challenges which require new methodologies to address, such as highly-siloed data, class imbalance, missing data, distribution shifts and non-standardised variables. Federated learning adds significant methodological complexity to conventional centralised machine learning, requiring distributed optimisation, communication between nodes, aggregation of models and redistribution of models. In this systematic review, we consider all papers on Scopus that were published between January 2015 and February 2023 and which describe new federated learning methodologies for addressing challenges with healthcare data. We performed a detailed review of the 89 papers which fulfilled these criteria. Significant systemic issues were identified throughout the literature which compromise the methodologies in many of the papers reviewed. We give detailed recommendations to help improve the quality of the methodology development for federated learning in healthcare.
REFORMS: Reporting Standards for Machine Learning Based Science
Aug 15, 2023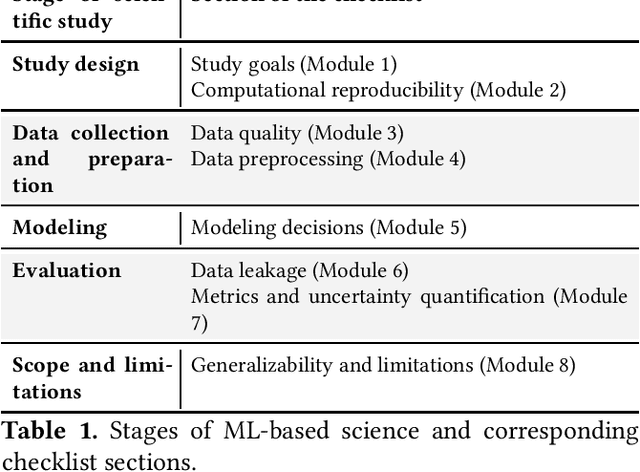
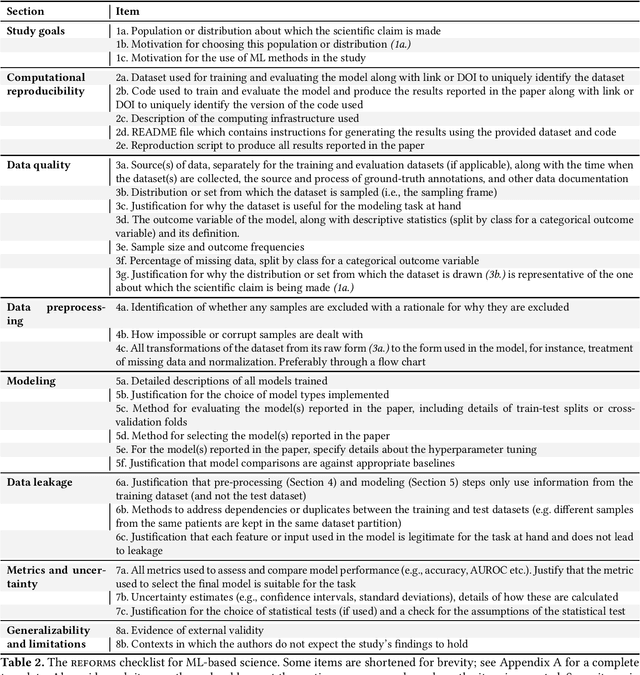
Abstract:Machine learning (ML) methods are proliferating in scientific research. However, the adoption of these methods has been accompanied by failures of validity, reproducibility, and generalizability. These failures can hinder scientific progress, lead to false consensus around invalid claims, and undermine the credibility of ML-based science. ML methods are often applied and fail in similar ways across disciplines. Motivated by this observation, our goal is to provide clear reporting standards for ML-based science. Drawing from an extensive review of past literature, we present the REFORMS checklist ($\textbf{Re}$porting Standards $\textbf{For}$ $\textbf{M}$achine Learning Based $\textbf{S}$cience). It consists of 32 questions and a paired set of guidelines. REFORMS was developed based on a consensus of 19 researchers across computer science, data science, mathematics, social sciences, and biomedical sciences. REFORMS can serve as a resource for researchers when designing and implementing a study, for referees when reviewing papers, and for journals when enforcing standards for transparency and reproducibility.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge