Janek Gröhl
Towards robust quantitative photoacoustic tomography via learned iterative methods
Oct 31, 2025Abstract:Photoacoustic tomography (PAT) is a medical imaging modality that can provide high-resolution tissue images based on the optical absorption. Classical reconstruction methods for quantifying the absorption coefficients rely on sufficient prior information to overcome noisy and imperfect measurements. As these methods utilize computationally expensive forward models, the computation becomes slow, limiting their potential for time-critical applications. As an alternative approach, deep learning-based reconstruction methods have been established for faster and more accurate reconstructions. However, most of these methods rely on having a large amount of training data, which is not the case in practice. In this work, we adopt the model-based learned iterative approach for the use in Quantitative PAT (QPAT), in which additional information from the model is iteratively provided to the updating networks, allowing better generalizability with scarce training data. We compare the performance of different learned updates based on gradient descent, Gauss-Newton, and Quasi-Newton methods. The learning tasks are formulated as greedy, requiring iterate-wise optimality, as well as end-to-end, where all networks are trained jointly. The implemented methods are tested with ideal simulated data as well as against a digital twin dataset that emulates scarce training data and high modeling error.
Digital twins enable full-reference quality assessment of photoacoustic image reconstructions
May 30, 2025



Abstract:Quantitative comparison of the quality of photoacoustic image reconstruction algorithms remains a major challenge. No-reference image quality measures are often inadequate, but full-reference measures require access to an ideal reference image. While the ground truth is known in simulations, it is unknown in vivo, or in phantom studies, as the reference depends on both the phantom properties and the imaging system. We tackle this problem by using numerical digital twins of tissue-mimicking phantoms and the imaging system to perform a quantitative calibration to reduce the simulation gap. The contributions of this paper are two-fold: First, we use this digital-twin framework to compare multiple state-of-the-art reconstruction algorithms. Second, among these is a Fourier transform-based reconstruction algorithm for circular detection geometries, which we test on experimental data for the first time. Our results demonstrate the usefulness of digital phantom twins by enabling assessment of the accuracy of the numerical forward model and enabling comparison of image reconstruction schemes with full-reference image quality assessment. We show that the Fourier transform-based algorithm yields results comparable to those of iterative time reversal, but at a lower computational cost. All data and code are publicly available on Zenodo: https://doi.org/10.5281/zenodo.15388429.
Parameter choices in HaarPSI for IQA with medical images
Oct 31, 2024Abstract:When developing machine learning models, image quality assessment (IQA) measures are a crucial component for evaluation. However, commonly used IQA measures have been primarily developed and optimized for natural images. In many specialized settings, such as medical images, this poses an often-overlooked problem regarding suitability. In previous studies, the IQA measure HaarPSI showed promising behavior for natural and medical images. HaarPSI is based on Haar wavelet representations and the framework allows optimization of two parameters. So far, these parameters have been aligned for natural images. Here, we optimize these parameters for two annotated medical data sets, a photoacoustic and a chest X-Ray data set. We observe that they are more sensitive to the parameter choices than the employed natural images, and on the other hand both medical data sets lead to similar parameter values when optimized. We denote the optimized setting, which improves the performance for the medical images notably, by HaarPSI$_{MED}$. The results suggest that adapting common IQA measures within their frameworks for medical images can provide a valuable, generalizable addition to the employment of more specific task-based measures.
A study on the adequacy of common IQA measures for medical images
May 29, 2024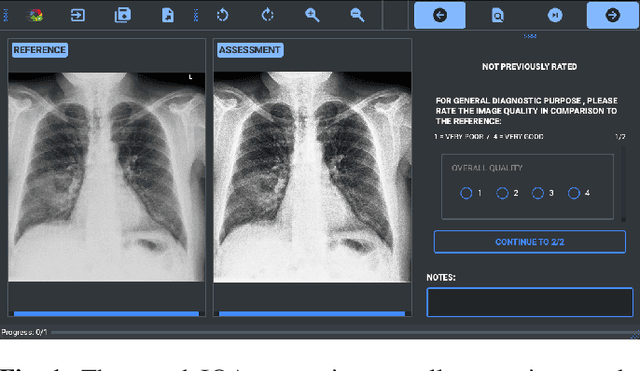
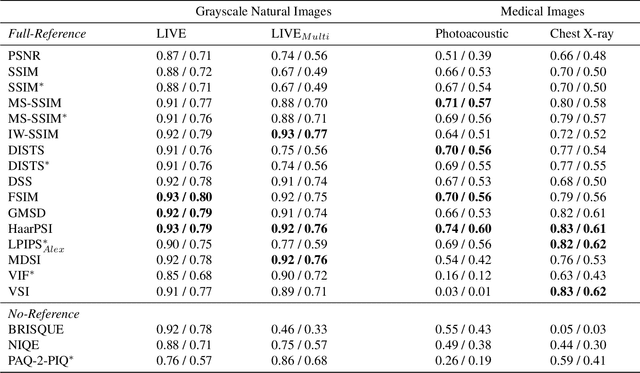
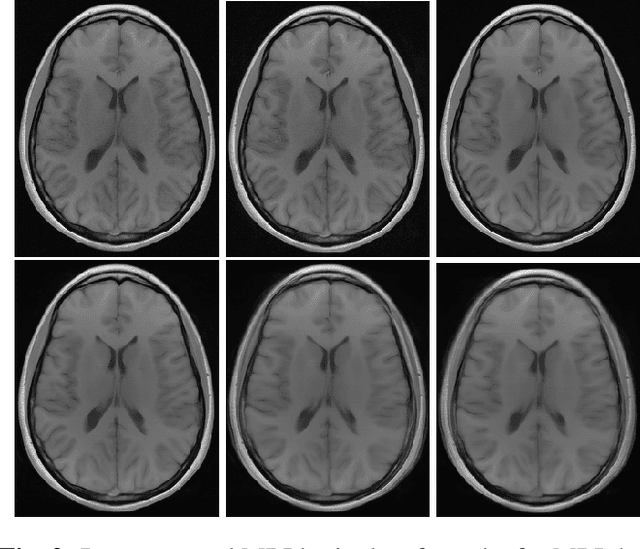
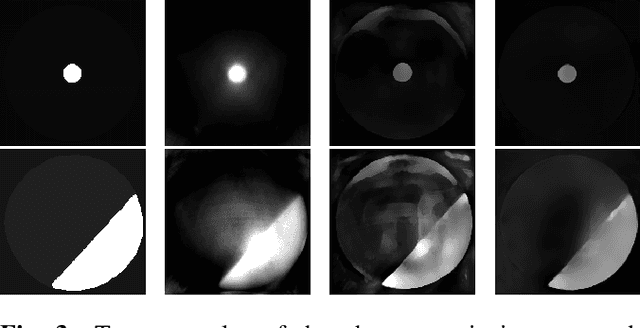
Abstract:Image quality assessment (IQA) is standard practice in the development stage of novel machine learning algorithms that operate on images. The most commonly used IQA measures have been developed and tested for natural images, but not in the medical setting. Reported inconsistencies arising in medical images are not surprising, as they have different properties than natural images. In this study, we test the applicability of common IQA measures for medical image data by comparing their assessment to manually rated chest X-ray (5 experts) and photoacoustic image data (1 expert). Moreover, we include supplementary studies on grayscale natural images and accelerated brain MRI data. The results of all experiments show a similar outcome in line with previous findings for medical imaging: PSNR and SSIM in the default setting are in the lower range of the result list and HaarPSI outperforms the other tested measures in the overall performance. Also among the top performers in our medical experiments are the full reference measures DISTS, FSIM, LPIPS and MS-SSIM. Generally, the results on natural images yield considerably higher correlations, suggesting that the additional employment of tailored IQA measures for medical imaging algorithms is needed.
A study of why we need to reassess full reference image quality assessment with medical images
May 29, 2024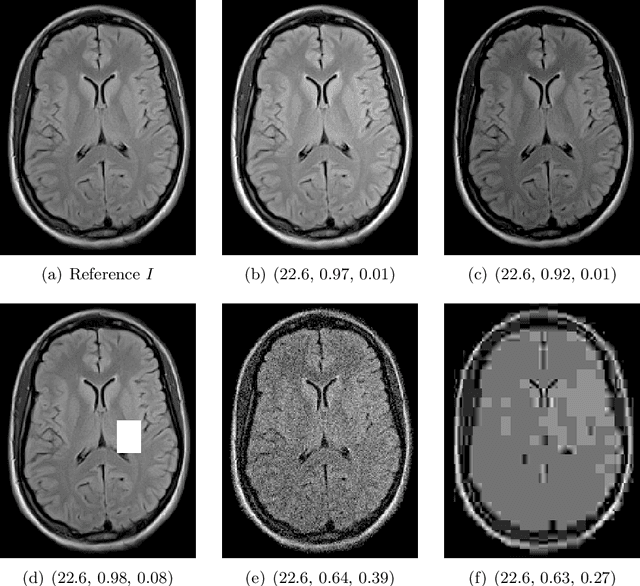
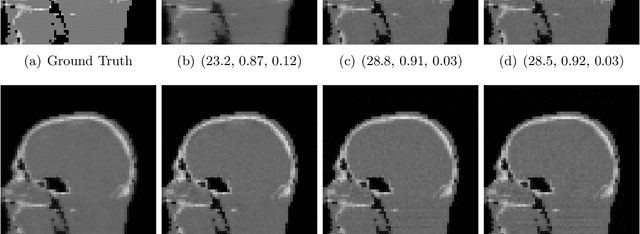
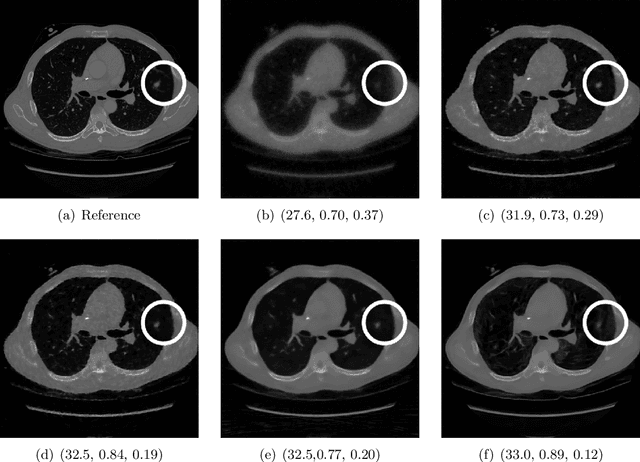
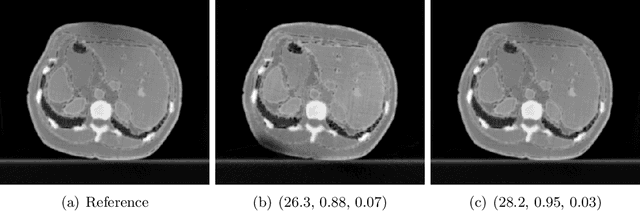
Abstract:Image quality assessment (IQA) is not just indispensable in clinical practice to ensure high standards, but also in the development stage of novel algorithms that operate on medical images with reference data. This paper provides a structured and comprehensive collection of examples where the two most common full reference (FR) image quality measures prove to be unsuitable for the assessment of novel algorithms using different kinds of medical images, including real-world MRI, CT, OCT, X-Ray, digital pathology and photoacoustic imaging data. In particular, the FR-IQA measures PSNR and SSIM are known and tested for working successfully in many natural imaging tasks, but discrepancies in medical scenarios have been noted in the literature. Inconsistencies arising in medical images are not surprising, as they have very different properties than natural images which have not been targeted nor tested in the development of the mentioned measures, and therefore might imply wrong judgement of novel methods for medical images. Therefore, improvement is urgently needed in particular in this era of AI to increase explainability, reproducibility and generalizability in machine learning for medical imaging and beyond. On top of the pitfalls we will provide ideas for future research as well as suggesting guidelines for the usage of FR-IQA measures applied to medical images.
Distribution-informed and wavelength-flexible data-driven photoacoustic oximetry
Mar 21, 2024Abstract:Significance: Photoacoustic imaging (PAI) promises to measure spatially-resolved blood oxygen saturation, but suffers from a lack of accurate and robust spectral unmixing methods to deliver on this promise. Accurate blood oxygenation estimation could have important clinical applications, from cancer detection to quantifying inflammation. Aim: This study addresses the inflexibility of existing data-driven methods for estimating blood oxygenation in PAI by introducing a recurrent neural network architecture. Approach: We created 25 simulated training dataset variations to assess neural network performance. We used a long short-term memory network to implement a wavelength-flexible network architecture and proposed the Jensen-Shannon divergence to predict the most suitable training dataset. Results: The network architecture can handle arbitrary input wavelengths and outperforms linear unmixing and the previously proposed learned spectral decolouring method. Small changes in the training data significantly affect the accuracy of our method, but we find that the Jensen-Shannon divergence correlates with the estimation error and is thus suitable for predicting the most appropriate training datasets for any given application. Conclusions: A flexible data-driven network architecture combined with the Jensen-Shannon Divergence to predict the best training data set provides a promising direction that might enable robust data-driven photoacoustic oximetry for clinical use cases.
Moving beyond simulation: data-driven quantitative photoacoustic imaging using tissue-mimicking phantoms
Jun 11, 2023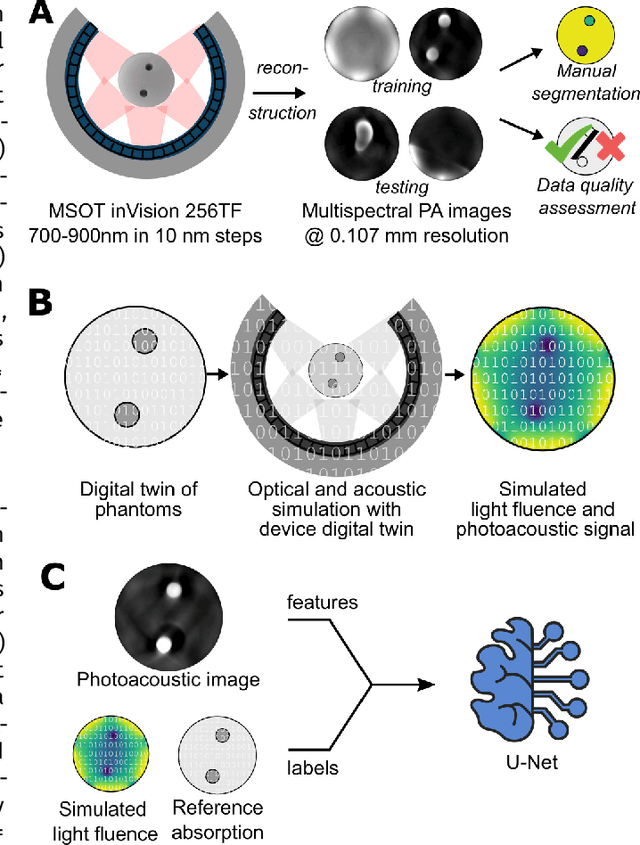
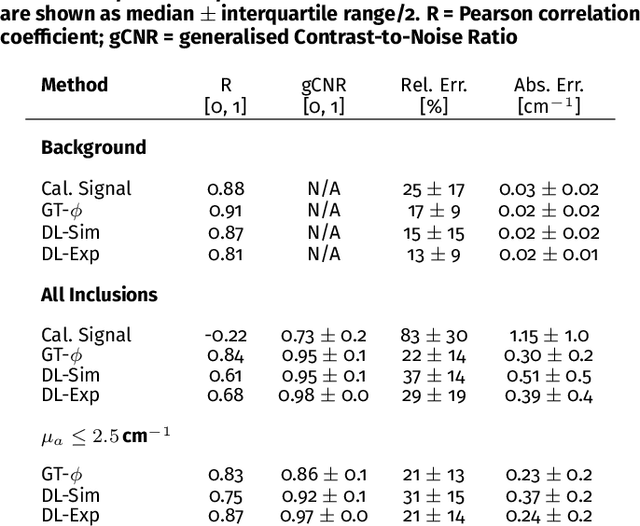
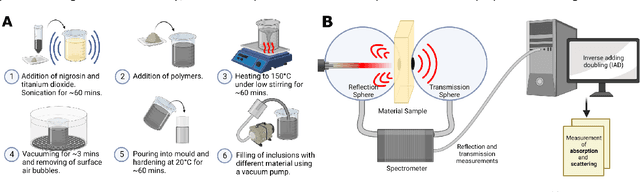
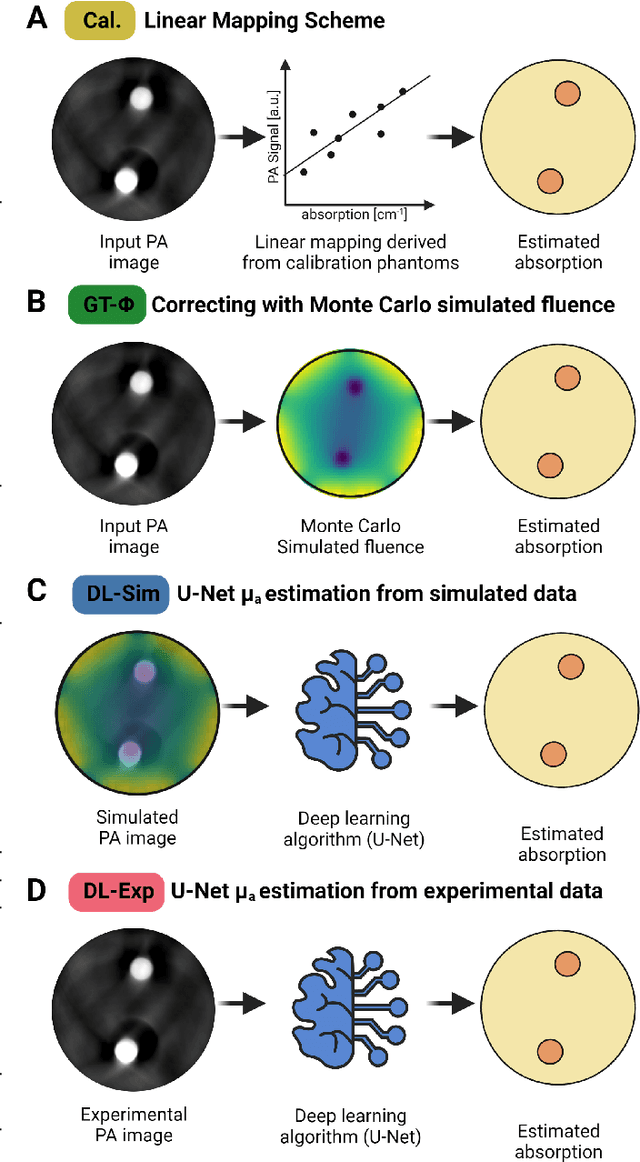
Abstract:Accurate measurement of optical absorption coefficients from photoacoustic imaging (PAI) data would enable direct mapping of molecular concentrations, providing vital clinical insight. The ill-posed nature of the problem of absorption coefficient recovery has prohibited PAI from achieving this goal in living systems due to the domain gap between simulation and experiment. To bridge this gap, we introduce a collection of experimentally well-characterised imaging phantoms and their digital twins. This first-of-a-kind phantom data set enables supervised training of a U-Net on experimental data for pixel-wise estimation of absorption coefficients. We show that training on simulated data results in artefacts and biases in the estimates, reinforcing the existence of a domain gap between simulation and experiment. Training on experimentally acquired data, however, yielded more accurate and robust estimates of optical absorption coefficients. We compare the results to fluence correction with a Monte Carlo model from reference optical properties of the materials, which yields a quantification error of approximately 20%. Application of the trained U-Nets to a blood flow phantom demonstrated spectral biases when training on simulated data, while application to a mouse model highlighted the ability of both learning-based approaches to recover the depth-dependent loss of signal intensity. We demonstrate that training on experimental phantoms can restore the correlation of signal amplitudes measured in depth. While the absolute quantification error remains high and further improvements are needed, our results highlight the promise of deep learning to advance quantitative PAI.
Unsupervised Domain Transfer with Conditional Invertible Neural Networks
Mar 17, 2023Abstract:Synthetic medical image generation has evolved as a key technique for neural network training and validation. A core challenge, however, remains in the domain gap between simulations and real data. While deep learning-based domain transfer using Cycle Generative Adversarial Networks and similar architectures has led to substantial progress in the field, there are use cases in which state-of-the-art approaches still fail to generate training images that produce convincing results on relevant downstream tasks. Here, we address this issue with a domain transfer approach based on conditional invertible neural networks (cINNs). As a particular advantage, our method inherently guarantees cycle consistency through its invertible architecture, and network training can efficiently be conducted with maximum likelihood training. To showcase our method's generic applicability, we apply it to two spectral imaging modalities at different scales, namely hyperspectral imaging (pixel-level) and photoacoustic tomography (image-level). According to comprehensive experiments, our method enables the generation of realistic spectral data and outperforms the state of the art on two downstream classification tasks (binary and multi-class). cINN-based domain transfer could thus evolve as an important method for realistic synthetic data generation in the field of spectral imaging and beyond.
Semantic segmentation of multispectral photoacoustic images using deep learning
May 20, 2021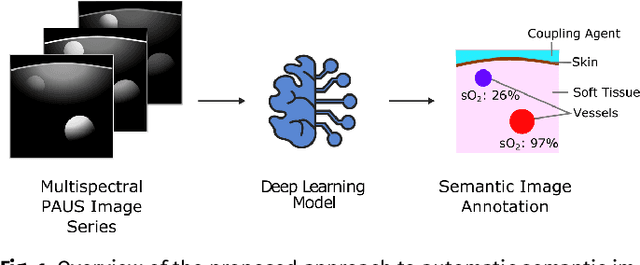
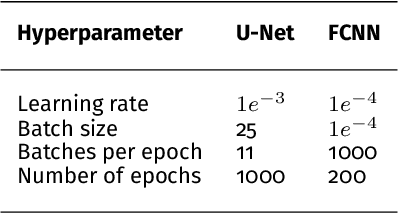
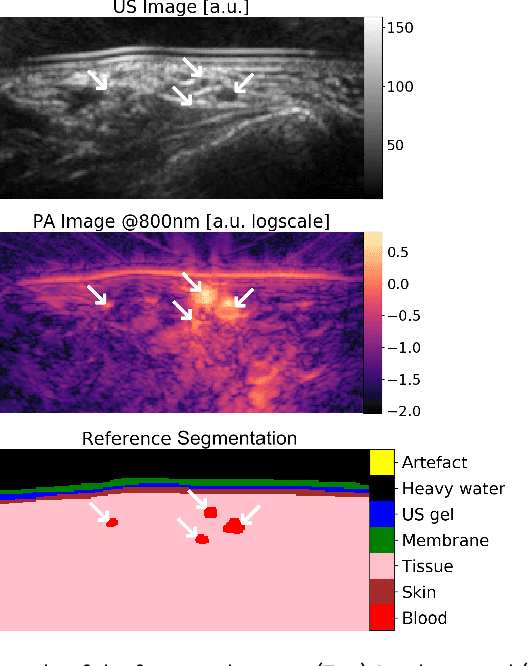
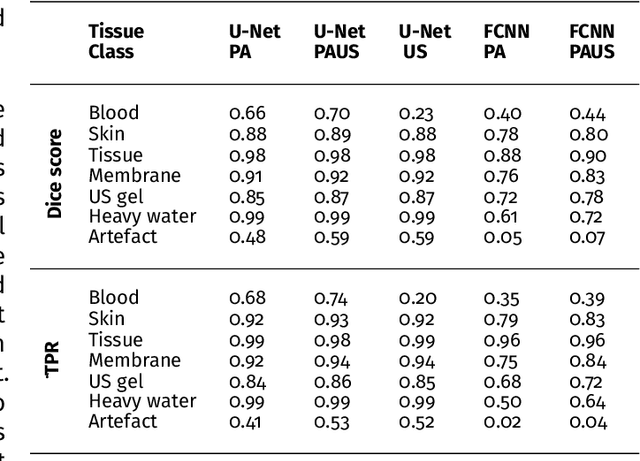
Abstract:Photoacoustic imaging has the potential to revolutionise healthcare due to the valuable information on tissue physiology that is contained in multispectral photoacoustic measurements. Clinical translation of the technology requires conversion of the high-dimensional acquired data into clinically relevant and interpretable information. In this work, we present a deep learning-based approach to semantic segmentation of multispectral photoacoustic images to facilitate the interpretability of recorded images. Manually annotated multispectral photoacoustic imaging data are used as gold standard reference annotations and enable the training of a deep learning-based segmentation algorithm in a supervised manner. Based on a validation study with experimentally acquired data of healthy human volunteers, we show that automatic tissue segmentation can be used to create powerful analyses and visualisations of multispectral photoacoustic images. Due to the intuitive representation of high-dimensional information, such a processing algorithm could be a valuable means to facilitate the clinical translation of photoacoustic imaging.
Data-driven generation of plausible tissue geometries for realistic photoacoustic image synthesis
Mar 29, 2021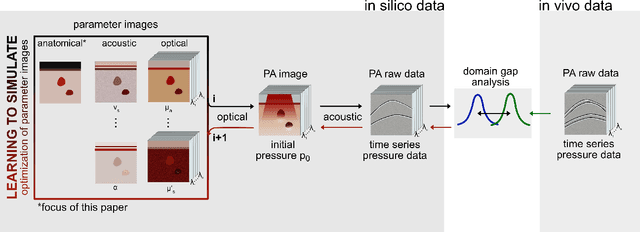
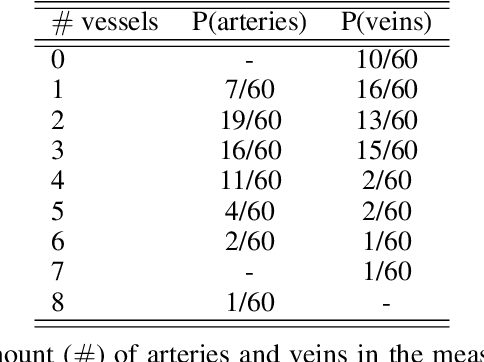
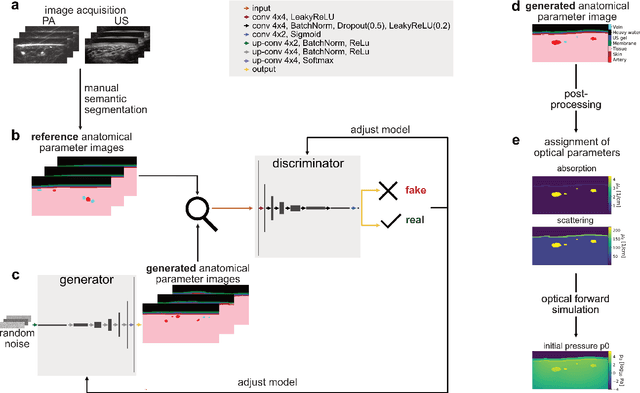
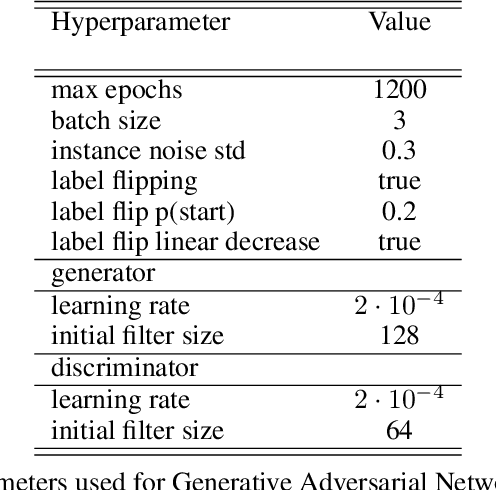
Abstract:Photoacoustic tomography (PAT) has the potential to recover morphological and functional tissue properties such as blood oxygenation with high spatial resolution and in an interventional setting. However, decades of research invested in solving the inverse problem of recovering clinically relevant tissue properties from spectral measurements have failed to produce solutions that can quantify tissue parameters robustly in a clinical setting. Previous attempts to address the limitations of model-based approaches with machine learning were hampered by the absence of labeled reference data needed for supervised algorithm training. While this bottleneck has been tackled by simulating training data, the domain gap between real and simulated images remains a huge unsolved challenge. As a first step to address this bottleneck, we propose a novel approach to PAT data simulation, which we refer to as "learning to simulate". Our approach involves subdividing the challenge of generating plausible simulations into two disjoint problems: (1) Probabilistic generation of realistic tissue morphology, represented by semantic segmentation maps and (2) pixel-wise assignment of corresponding optical and acoustic properties. In the present work, we focus on the first challenge. Specifically, we leverage the concept of Generative Adversarial Networks (GANs) trained on semantically annotated medical imaging data to generate plausible tissue geometries. According to an initial in silico feasibility study our approach is well-suited for contributing to realistic PAT image synthesis and could thus become a fundamental step for deep learning-based quantitative PAT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge