Kris Dreher
Semantic segmentation of multispectral photoacoustic images using deep learning
May 20, 2021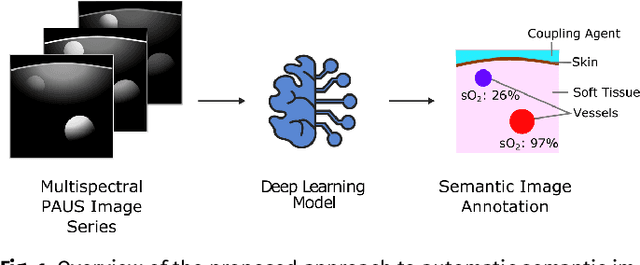
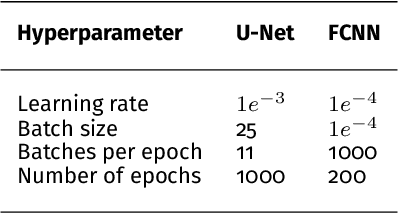
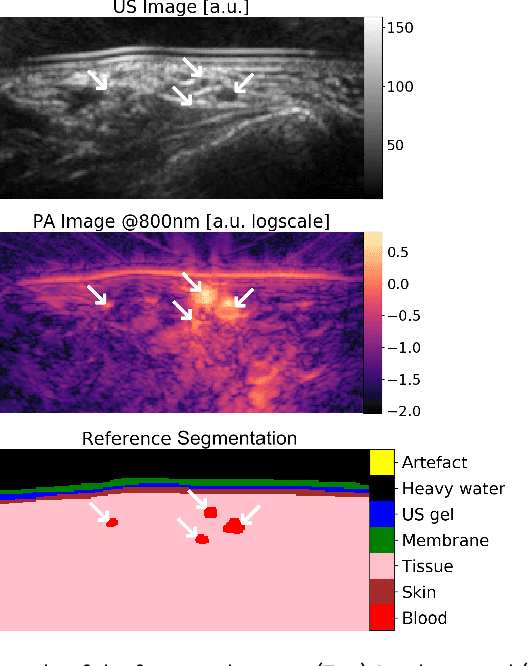
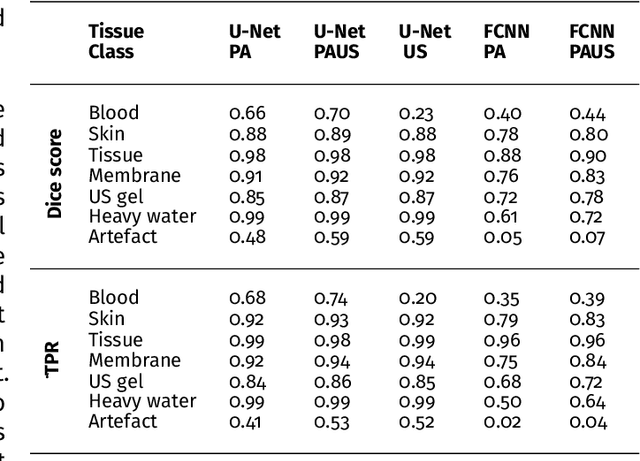
Abstract:Photoacoustic imaging has the potential to revolutionise healthcare due to the valuable information on tissue physiology that is contained in multispectral photoacoustic measurements. Clinical translation of the technology requires conversion of the high-dimensional acquired data into clinically relevant and interpretable information. In this work, we present a deep learning-based approach to semantic segmentation of multispectral photoacoustic images to facilitate the interpretability of recorded images. Manually annotated multispectral photoacoustic imaging data are used as gold standard reference annotations and enable the training of a deep learning-based segmentation algorithm in a supervised manner. Based on a validation study with experimentally acquired data of healthy human volunteers, we show that automatic tissue segmentation can be used to create powerful analyses and visualisations of multispectral photoacoustic images. Due to the intuitive representation of high-dimensional information, such a processing algorithm could be a valuable means to facilitate the clinical translation of photoacoustic imaging.
Data-driven generation of plausible tissue geometries for realistic photoacoustic image synthesis
Mar 29, 2021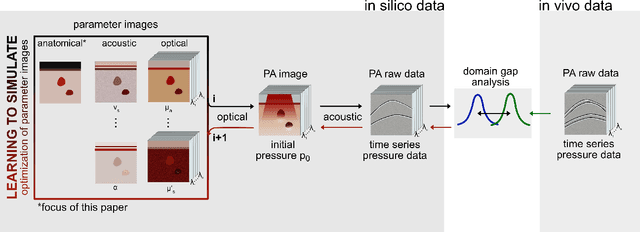
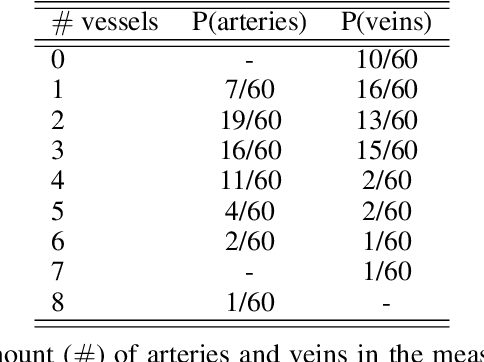
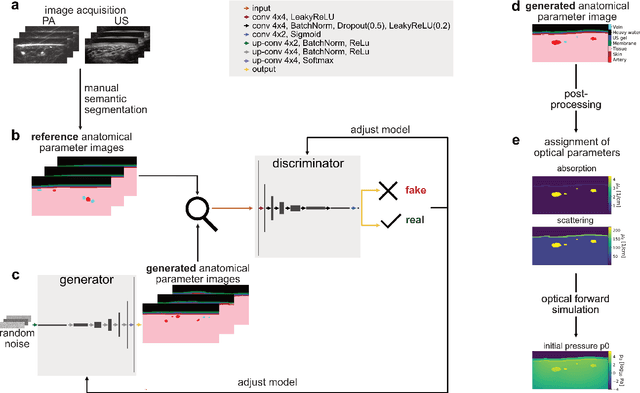
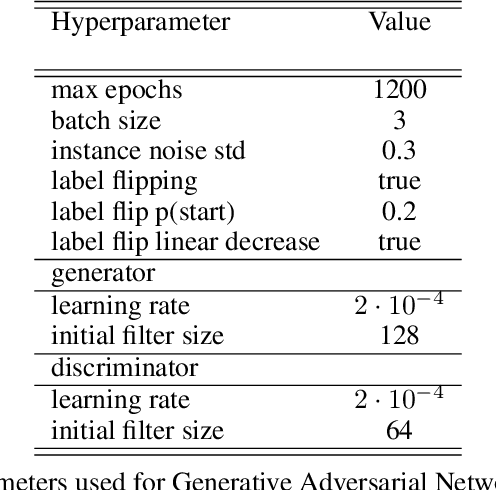
Abstract:Photoacoustic tomography (PAT) has the potential to recover morphological and functional tissue properties such as blood oxygenation with high spatial resolution and in an interventional setting. However, decades of research invested in solving the inverse problem of recovering clinically relevant tissue properties from spectral measurements have failed to produce solutions that can quantify tissue parameters robustly in a clinical setting. Previous attempts to address the limitations of model-based approaches with machine learning were hampered by the absence of labeled reference data needed for supervised algorithm training. While this bottleneck has been tackled by simulating training data, the domain gap between real and simulated images remains a huge unsolved challenge. As a first step to address this bottleneck, we propose a novel approach to PAT data simulation, which we refer to as "learning to simulate". Our approach involves subdividing the challenge of generating plausible simulations into two disjoint problems: (1) Probabilistic generation of realistic tissue morphology, represented by semantic segmentation maps and (2) pixel-wise assignment of corresponding optical and acoustic properties. In the present work, we focus on the first challenge. Specifically, we leverage the concept of Generative Adversarial Networks (GANs) trained on semantically annotated medical imaging data to generate plausible tissue geometries. According to an initial in silico feasibility study our approach is well-suited for contributing to realistic PAT image synthesis and could thus become a fundamental step for deep learning-based quantitative PAT.
Tattoo tomography: Freehand 3D photoacoustic image reconstruction with an optical pattern
Nov 11, 2020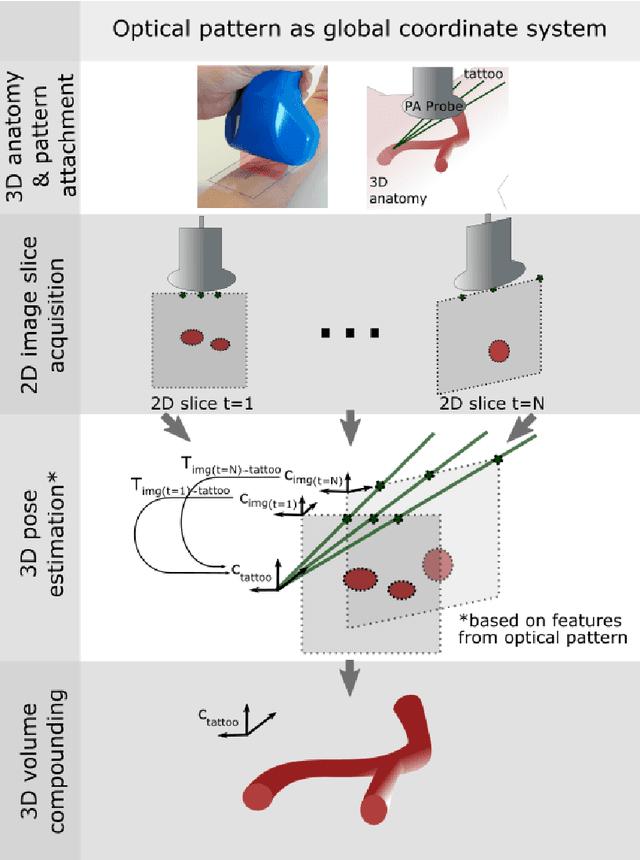
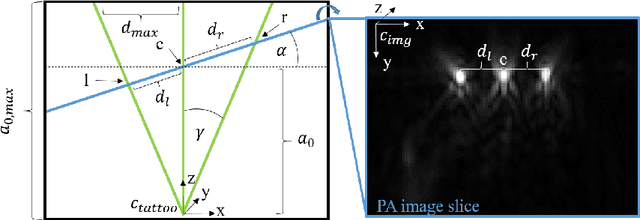
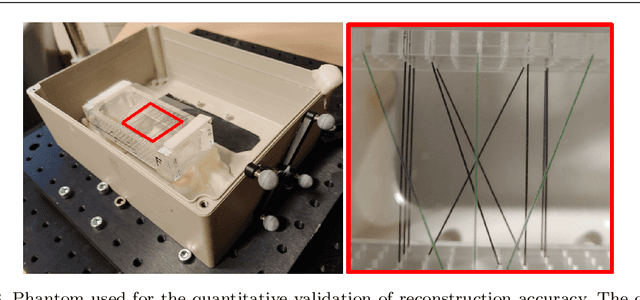

Abstract:Purpose: Photoacoustic tomography (PAT) is a novel imaging technique that can spatially resolve both morphological and functional tissue properties, such as the vessel topology and tissue oxygenation. While this capacity makes PAT a promising modality for the diagnosis, treatment and follow-up of various diseases, a current drawback is the limited field-of-view (FoV) provided by the conventionally applied 2D probes. Methods: In this paper, we present a novel approach to 3D reconstruction of PAT data (Tattoo tomography) that does not require an external tracking system and can smoothly be integrated into clinical workflows. It is based on an optical pattern placed on the region of interest prior to image acquisition. This pattern is designed in a way that a tomographic image of it enables the recovery of the probe pose relative to the coordinate system of the pattern. This allows the transformation of a sequence of acquired PA images into one common global coordinate system and thus the consistent 3D reconstruction of PAT imaging data. Results: An initial feasibility study conducted with experimental phantom data and in vivo forearm data indicates that the Tattoo approach is well-suited for 3D reconstruction of PAT data with high accuracy and precision. Conclusion: In contrast to previous approaches to 3D ultrasound (US) or PAT reconstruction, the Tattoo approach neither requires complex external hardware nor training data acquired for a specific application. It could thus become a valuable tool for clinical freehand PAT.
Deep learning for biomedical photoacoustic imaging: A review
Nov 05, 2020



Abstract:Photoacoustic imaging (PAI) is a promising emerging imaging modality that enables spatially resolved imaging of optical tissue properties up to several centimeters deep in tissue, creating the potential for numerous exciting clinical applications. However, extraction of relevant tissue parameters from the raw data requires the solving of inverse image reconstruction problems, which have proven extremely difficult to solve. The application of deep learning methods has recently exploded in popularity, leading to impressive successes in the context of medical imaging and also finding first use in the field of PAI. Deep learning methods possess unique advantages that can facilitate the clinical translation of PAI, such as extremely fast computation times and the fact that they can be adapted to any given problem. In this review, we examine the current state of the art regarding deep learning in PAI and identify potential directions of research that will help to reach the goal of clinical applicability
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge