Silvia Seidlitz
Computer Assisted Medical Interventions, Helmholtz Information and Data Science School for Health, Karlsruhe/Heidelberg, Germany
CARL: Camera-Agnostic Representation Learning for Spectral Image Analysis
Apr 27, 2025Abstract:Spectral imaging offers promising applications across diverse domains, including medicine and urban scene understanding, and is already established as a critical modality in remote sensing. However, variability in channel dimensionality and captured wavelengths among spectral cameras impede the development of AI-driven methodologies, leading to camera-specific models with limited generalizability and inadequate cross-camera applicability. To address this bottleneck, we introduce $\textbf{CARL}$, a model for $\textbf{C}$amera-$\textbf{A}$gnostic $\textbf{R}$epresentation $\textbf{L}$earning across RGB, multispectral, and hyperspectral imaging modalities. To enable the conversion of a spectral image with any channel dimensionality to a camera-agnostic embedding, we introduce wavelength positional encoding and a self-attention-cross-attention mechanism to compress spectral information into learned query representations. Spectral-spatial pre-training is achieved with a novel spectral self-supervised JEPA-inspired strategy tailored to CARL. Large-scale experiments across the domains of medical imaging, autonomous driving, and satellite imaging demonstrate our model's unique robustness to spectral heterogeneity, outperforming on datasets with simulated and real-world cross-camera spectral variations. The scalability and versatility of the proposed approach position our model as a backbone for future spectral foundation models.
Deep intra-operative illumination calibration of hyperspectral cameras
Sep 11, 2024



Abstract:Hyperspectral imaging (HSI) is emerging as a promising novel imaging modality with various potential surgical applications. Currently available cameras, however, suffer from poor integration into the clinical workflow because they require the lights to be switched off, or the camera to be manually recalibrated as soon as lighting conditions change. Given this critical bottleneck, the contribution of this paper is threefold: (1) We demonstrate that dynamically changing lighting conditions in the operating room dramatically affect the performance of HSI applications, namely physiological parameter estimation, and surgical scene segmentation. (2) We propose a novel learning-based approach to automatically recalibrating hyperspectral images during surgery and show that it is sufficiently accurate to replace the tedious process of white reference-based recalibration. (3) Based on a total of 742 HSI cubes from a phantom, porcine models, and rats we show that our recalibration method not only outperforms previously proposed methods, but also generalizes across species, lighting conditions, and image processing tasks. Due to its simple workflow integration as well as high accuracy, speed, and generalization capabilities, our method could evolve as a central component in clinical surgical HSI.
Handling Geometric Domain Shifts in Semantic Segmentation of Surgical RGB and Hyperspectral Images
Aug 27, 2024



Abstract:Robust semantic segmentation of intraoperative image data holds promise for enabling automatic surgical scene understanding and autonomous robotic surgery. While model development and validation are primarily conducted on idealistic scenes, geometric domain shifts, such as occlusions of the situs, are common in real-world open surgeries. To close this gap, we (1) present the first analysis of state-of-the-art (SOA) semantic segmentation models when faced with geometric out-of-distribution (OOD) data, and (2) propose an augmentation technique called "Organ Transplantation", to enhance generalizability. Our comprehensive validation on six different OOD datasets, comprising 600 RGB and hyperspectral imaging (HSI) cubes from 33 pigs, each annotated with 19 classes, reveals a large performance drop in SOA organ segmentation models on geometric OOD data. This performance decline is observed not only in conventional RGB data (with a dice similarity coefficient (DSC) drop of 46 %) but also in HSI data (with a DSC drop of 45 %), despite the richer spectral information content. The performance decline increases with the spatial granularity of the input data. Our augmentation technique improves SOA model performance by up to 67 % for RGB data and 90 % for HSI data, achieving performance at the level of in-distribution performance on real OOD test data. Given the simplicity and effectiveness of our augmentation method, it is a valuable tool for addressing geometric domain shifts in surgical scene segmentation, regardless of the underlying model. Our code and pre-trained models are publicly available at https://github.com/IMSY-DKFZ/htc.
New spectral imaging biomarkers for sepsis and mortality in intensive care
Aug 19, 2024Abstract:With sepsis remaining a leading cause of mortality, early identification of septic patients and those at high risk of death is a challenge of high socioeconomic importance. The driving hypothesis of this study was that hyperspectral imaging (HSI) could provide novel biomarkers for sepsis diagnosis and treatment management due to its potential to monitor microcirculatory alterations. We conducted a comprehensive study involving HSI data of the palm and fingers from more than 480 patients on the day of their intensive care unit (ICU) admission. The findings demonstrate that HSI measurements can predict sepsis with an area under the receiver operating characteristic curve (AUROC) of 0.80 (95 % confidence interval (CI) [0.76; 0.84]) and mortality with an AUROC of 0.72 (95 % CI [0.65; 0.79]). The predictive performance improves substantially when additional clinical data is incorporated, leading to an AUROC of up to 0.94 (95 % CI [0.92; 0.96]) for sepsis and 0.84 (95 % CI [0.78; 0.89]) for mortality. We conclude that HSI presents novel imaging biomarkers for the rapid, non-invasive prediction of sepsis and mortality, suggesting its potential as an important modality for guiding diagnosis and treatment.
Semantic segmentation of surgical hyperspectral images under geometric domain shifts
Mar 20, 2023



Abstract:Robust semantic segmentation of intraoperative image data could pave the way for automatic surgical scene understanding and autonomous robotic surgery. Geometric domain shifts, however, although common in real-world open surgeries due to variations in surgical procedures or situs occlusions, remain a topic largely unaddressed in the field. To address this gap in the literature, we (1) present the first analysis of state-of-the-art (SOA) semantic segmentation networks in the presence of geometric out-of-distribution (OOD) data, and (2) address generalizability with a dedicated augmentation technique termed "Organ Transplantation" that we adapted from the general computer vision community. According to a comprehensive validation on six different OOD data sets comprising 600 RGB and hyperspectral imaging (HSI) cubes from 33 pigs semantically annotated with 19 classes, we demonstrate a large performance drop of SOA organ segmentation networks applied to geometric OOD data. Surprisingly, this holds true not only for conventional RGB data (drop of Dice similarity coefficient (DSC) by 46 %) but also for HSI data (drop by 45 %), despite the latter's rich information content per pixel. Using our augmentation scheme improves on the SOA DSC by up to 67 % (RGB) and 90 % (HSI) and renders performance on par with in-distribution performance on real OOD test data. The simplicity and effectiveness of our augmentation scheme makes it a valuable network-independent tool for addressing geometric domain shifts in semantic scene segmentation of intraoperative data. Our code and pre-trained models will be made publicly available.
Unsupervised Domain Transfer with Conditional Invertible Neural Networks
Mar 17, 2023Abstract:Synthetic medical image generation has evolved as a key technique for neural network training and validation. A core challenge, however, remains in the domain gap between simulations and real data. While deep learning-based domain transfer using Cycle Generative Adversarial Networks and similar architectures has led to substantial progress in the field, there are use cases in which state-of-the-art approaches still fail to generate training images that produce convincing results on relevant downstream tasks. Here, we address this issue with a domain transfer approach based on conditional invertible neural networks (cINNs). As a particular advantage, our method inherently guarantees cycle consistency through its invertible architecture, and network training can efficiently be conducted with maximum likelihood training. To showcase our method's generic applicability, we apply it to two spectral imaging modalities at different scales, namely hyperspectral imaging (pixel-level) and photoacoustic tomography (image-level). According to comprehensive experiments, our method enables the generation of realistic spectral data and outperforms the state of the art on two downstream classification tasks (binary and multi-class). cINN-based domain transfer could thus evolve as an important method for realistic synthetic data generation in the field of spectral imaging and beyond.
Robust deep learning-based semantic organ segmentation in hyperspectral images
Nov 09, 2021Abstract:Semantic image segmentation is an important prerequisite for context-awareness and autonomous robotics in surgery. The state of the art has focused on conventional RGB video data acquired during minimally invasive surgery, but full-scene semantic segmentation based on spectral imaging data and obtained during open surgery has received almost no attention to date. To address this gap in the literature, we are investigating the following research questions based on hyperspectral imaging (HSI) data of pigs acquired in an open surgery setting: (1) What is an adequate representation of HSI data for neural network-based fully automated organ segmentation, especially with respect to the spatial granularity of the data (pixels vs. superpixels vs. patches vs. full images)? (2) Is there a benefit of using HSI data compared to other modalities, namely RGB data and processed HSI data (e.g. tissue parameters like oxygenation), when performing semantic organ segmentation? According to a comprehensive validation study based on 506 HSI images from 20 pigs, annotated with a total of 19 classes, deep learning-based segmentation performance increases - consistently across modalities - with the spatial context of the input data. Unprocessed HSI data offers an advantage over RGB data or processed data from the camera provider, with the advantage increasing with decreasing size of the input to the neural network. Maximum performance (HSI applied to whole images) yielded a mean dice similarity coefficient (DSC) of 0.89 (standard deviation (SD) 0.04), which is in the range of the inter-rater variability (DSC of 0.89 (SD 0.07)). We conclude that HSI could become a powerful image modality for fully-automatic surgical scene understanding with many advantages over traditional imaging, including the ability to recover additional functional tissue information.
Machine learning-based analysis of hyperspectral images for automated sepsis diagnosis
Jun 15, 2021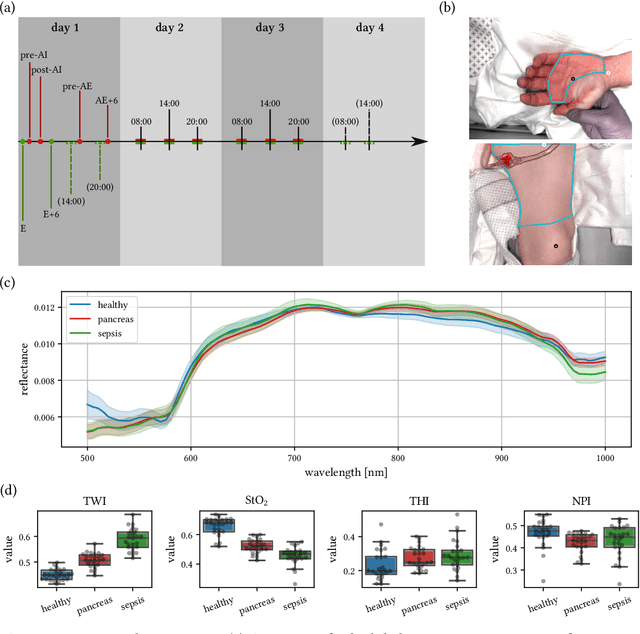
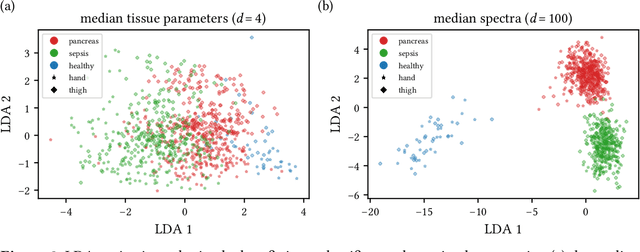
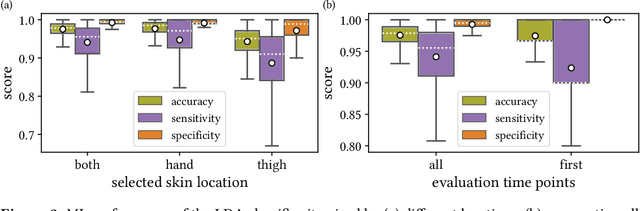
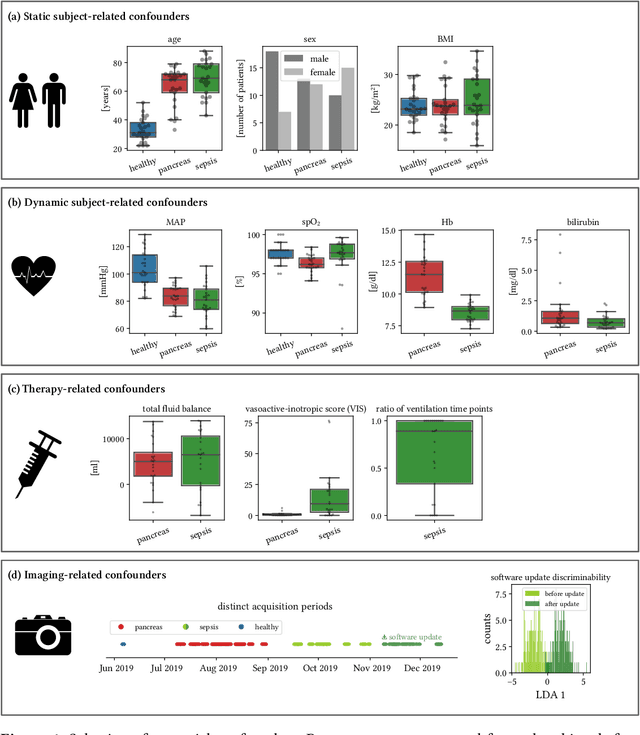
Abstract:Sepsis is a leading cause of mortality and critical illness worldwide. While robust biomarkers for early diagnosis are still missing, recent work indicates that hyperspectral imaging (HSI) has the potential to overcome this bottleneck by monitoring microcirculatory alterations. Automated machine learning-based diagnosis of sepsis based on HSI data, however, has not been explored to date. Given this gap in the literature, we leveraged an existing data set to (1) investigate whether HSI-based automated diagnosis of sepsis is possible and (2) put forth a list of possible confounders relevant for HSI-based tissue classification. While we were able to classify sepsis with an accuracy of over $98\,\%$ using the existing data, our research also revealed several subject-, therapy- and imaging-related confounders that may lead to an overestimation of algorithm performance when not balanced across the patient groups. We conclude that further prospective studies, carefully designed with respect to these confounders, are necessary to confirm the preliminary results obtained in this study.
Video-rate multispectral imaging in laparoscopic surgery: First-in-human application
May 28, 2021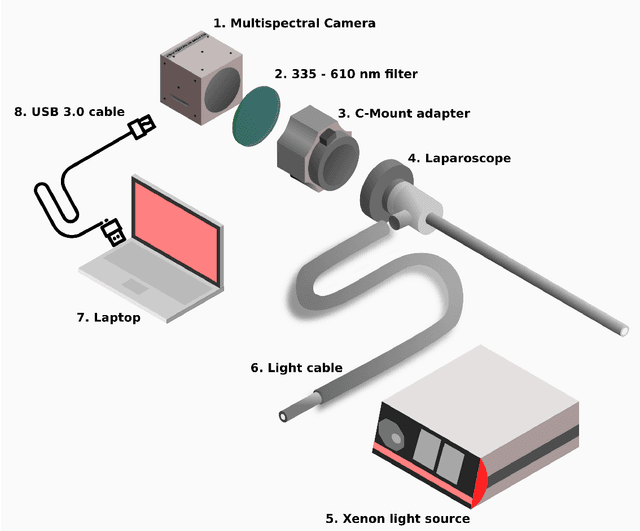
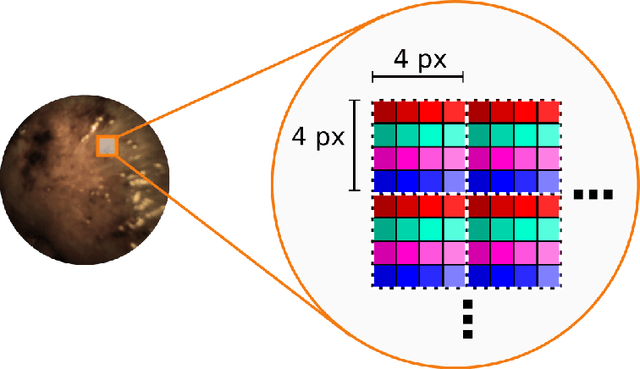
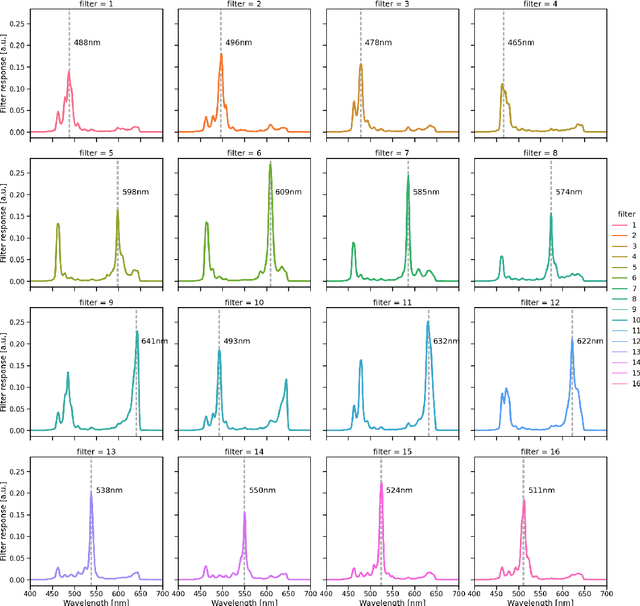
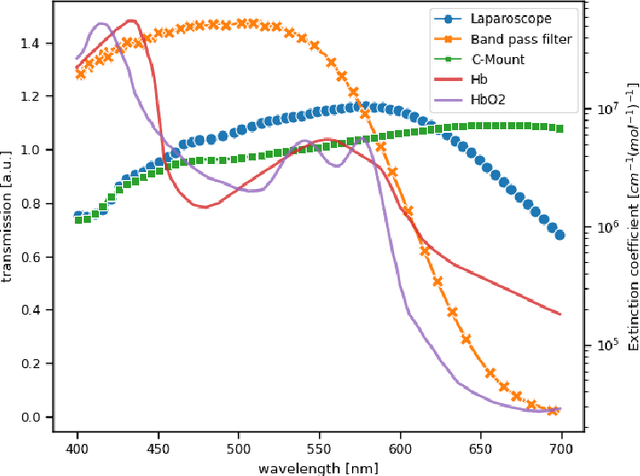
Abstract:Multispectral and hyperspectral imaging (MSI/HSI) can provide clinically relevant information on morphological and functional tissue properties. Application in the operating room (OR), however, has so far been limited by complex hardware setups and slow acquisition times. To overcome these limitations, we propose a novel imaging system for video-rate spectral imaging in the clinical workflow. The system integrates a small snapshot multispectral camera with a standard laparoscope and a clinically commonly used light source, enabling the recording of multispectral images with a spectral dimension of 16 at a frame rate of 25 Hz. An ongoing in patient study shows that multispectral recordings from this system can help detect perfusion changes in partial nephrectomy surgery, thus opening the doors to a wide range of clinical applications.
Heidelberg Colorectal Data Set for Surgical Data Science in the Sensor Operating Room
May 28, 2020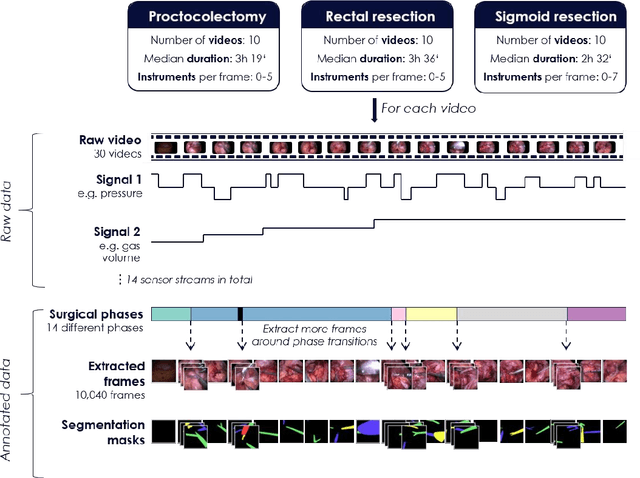
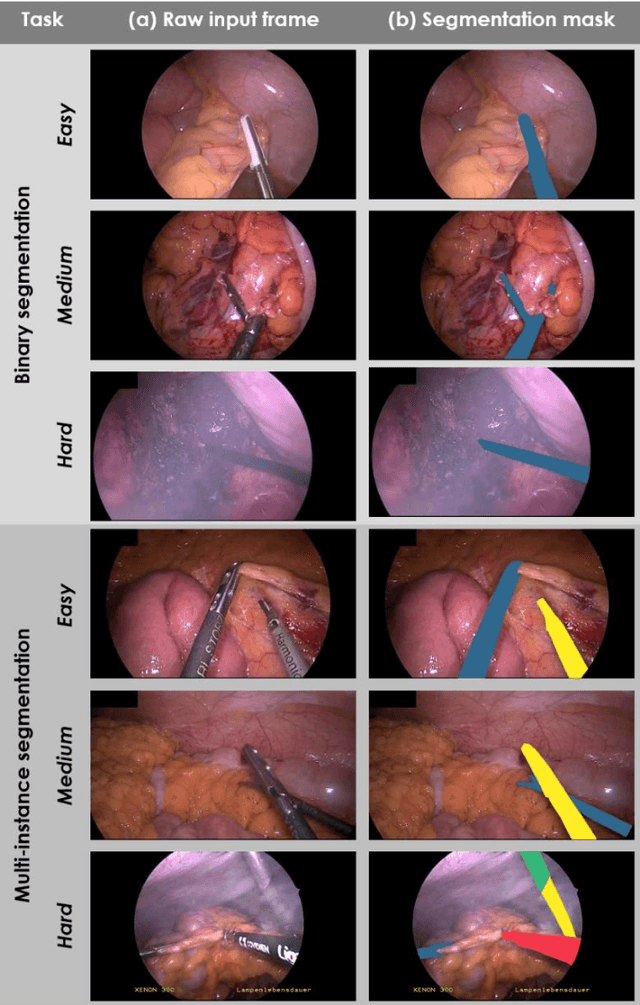
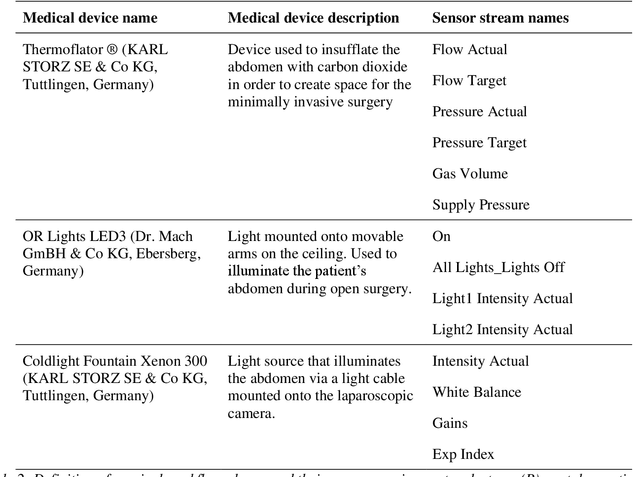
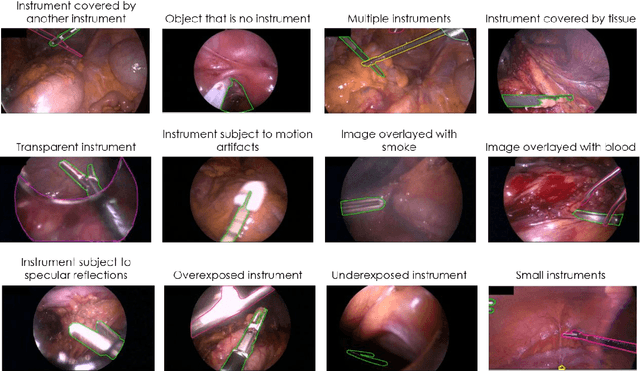
Abstract:Image-based tracking of medical instruments is an integral part of many surgical data science applications. Previous research has addressed the tasks of detecting, segmenting and tracking medical instruments based on laparoscopic video data. However, the methods proposed still tend to fail when applied to challenging images and do not generalize well to data they have not been trained on. This paper introduces the Heidelberg Colorectal (HeiCo) data set - the first publicly available data set enabling comprehensive benchmarking of medical instrument detection and segmentation algorithms with a specific emphasis on robustness and generalization capabilities of the methods. Our data set comprises 30 laparoscopic videos and corresponding sensor data from medical devices in the operating room for three different types of laparoscopic surgery. Annotations include surgical phase labels for all frames in the videos as well as instance-wise segmentation masks for surgical instruments in more than 10,000 individual frames. The data has successfully been used to organize international competitions in the scope of the Endoscopic Vision Challenges (EndoVis) 2017 and 2019.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge