Peter M. Full
*: shared first/last authors
Accurate Detection of Mediastinal Lesions with nnDetection
Mar 20, 2023Abstract:The accurate detection of mediastinal lesions is one of the rarely explored medical object detection problems. In this work, we applied a modified version of the self-configuring method nnDetection to the Mediastinal Lesion Analysis (MELA) Challenge 2022. By incorporating automatically generated pseudo masks, training high capacity models with large patch sizes in a multi GPU setup and an adapted augmentation scheme to reduce localization errors caused by rotations, our method achieved an excellent FROC score of 0.9922 at IoU 0.10 and 0.9880 at IoU 0.3 in our cross-validation experiments. The submitted ensemble ranked third in the competition with a FROC score of 0.9897 on the MELA challenge leaderboard.
How can we learn from challenges? A statistical approach to driving future algorithm development
Jun 17, 2021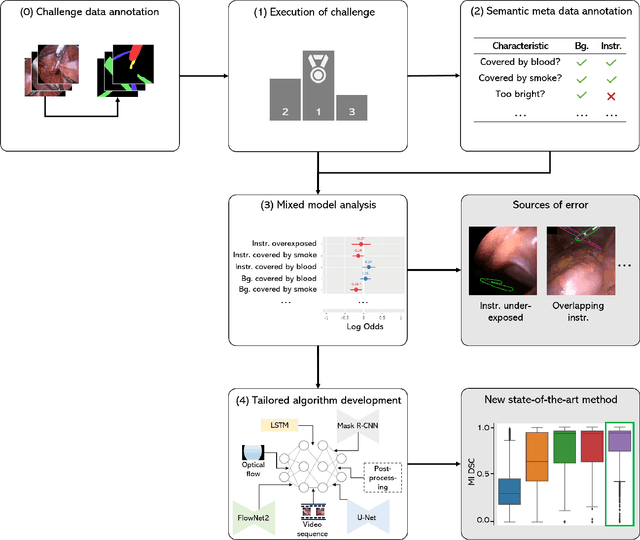
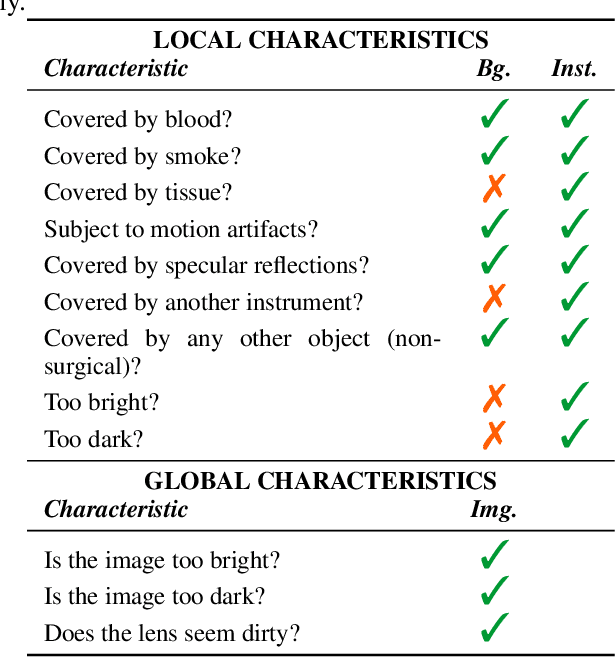
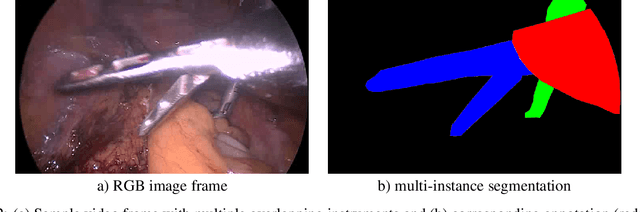
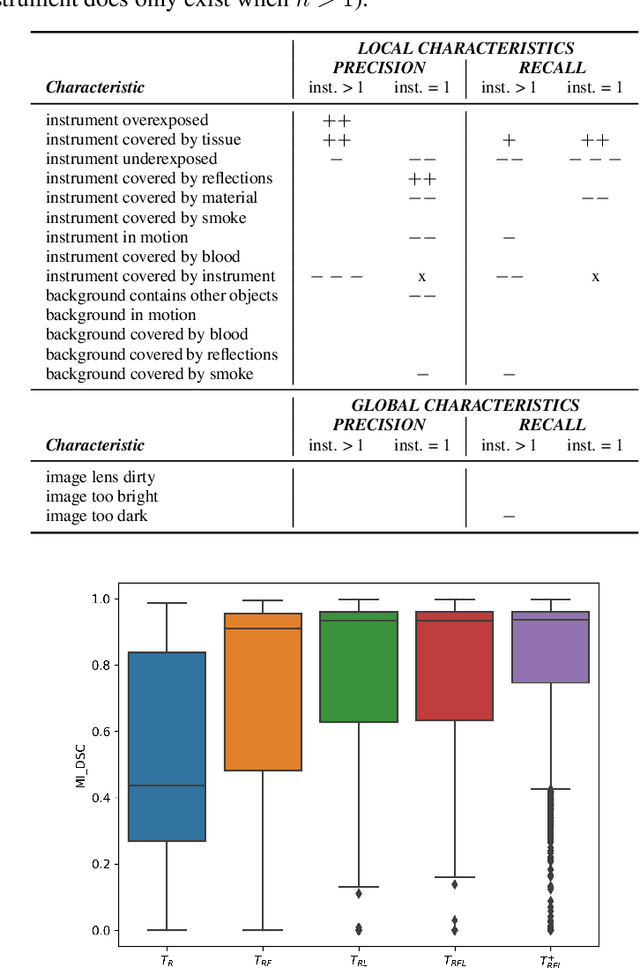
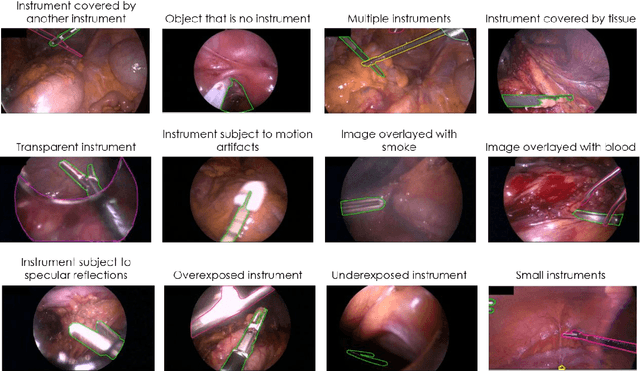
Abstract:Challenges have become the state-of-the-art approach to benchmark image analysis algorithms in a comparative manner. While the validation on identical data sets was a great step forward, results analysis is often restricted to pure ranking tables, leaving relevant questions unanswered. Specifically, little effort has been put into the systematic investigation on what characterizes images in which state-of-the-art algorithms fail. To address this gap in the literature, we (1) present a statistical framework for learning from challenges and (2) instantiate it for the specific task of instrument instance segmentation in laparoscopic videos. Our framework relies on the semantic meta data annotation of images, which serves as foundation for a General Linear Mixed Models (GLMM) analysis. Based on 51,542 meta data annotations performed on 2,728 images, we applied our approach to the results of the Robust Medical Instrument Segmentation Challenge (ROBUST-MIS) challenge 2019 and revealed underexposure, motion and occlusion of instruments as well as the presence of smoke or other objects in the background as major sources of algorithm failure. Our subsequent method development, tailored to the specific remaining issues, yielded a deep learning model with state-of-the-art overall performance and specific strengths in the processing of images in which previous methods tended to fail. Due to the objectivity and generic applicability of our approach, it could become a valuable tool for validation in the field of medical image analysis and beyond. and segmentation of small, crossing, moving and transparent instrument(s) (parts).
Studying Robustness of Semantic Segmentation under Domain Shift in cardiac MRI
Nov 15, 2020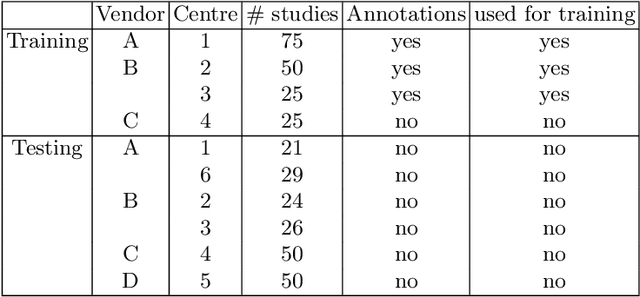
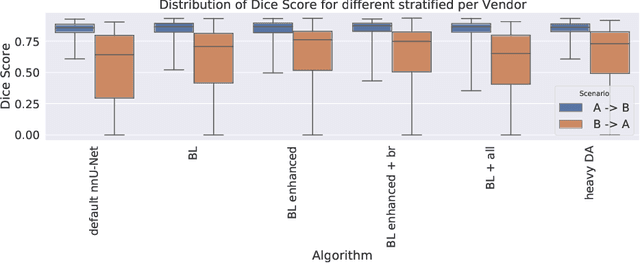

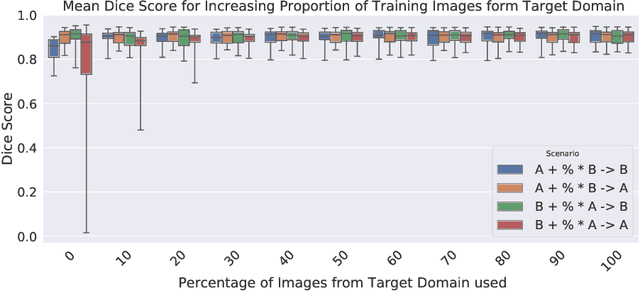
Abstract:Cardiac magnetic resonance imaging (cMRI) is an integral part of diagnosis in many heart related diseases. Recently, deep neural networks have demonstrated successful automatic segmentation, thus alleviating the burden of time-consuming manual contouring of cardiac structures. Moreover, frameworks such as nnU-Net provide entirely automatic model configuration to unseen datasets enabling out-of-the-box application even by non-experts. However, current studies commonly neglect the clinically realistic scenario, in which a trained network is applied to data from a different domain such as deviating scanners or imaging protocols. This potentially leads to unexpected performance drops of deep learning models in real life applications. In this work, we systematically study challenges and opportunities of domain transfer across images from multiple clinical centres and scanner vendors. In order to maintain out-of-the-box usability, we build upon a fixed U-Net architecture configured by the nnU-net framework to investigate various data augmentation techniques and batch normalization layers as an easy-to-customize pipeline component and provide general guidelines on how to improve domain generalizability abilities in existing deep learning methods. Our proposed method ranked first at the Multi-Centre, Multi-Vendor & Multi-Disease Cardiac Image Segmentation Challenge (M&Ms).
nnU-Net for Brain Tumor Segmentation
Nov 02, 2020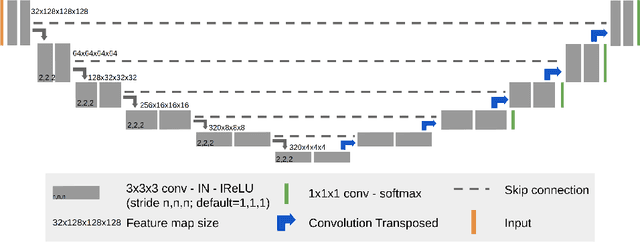
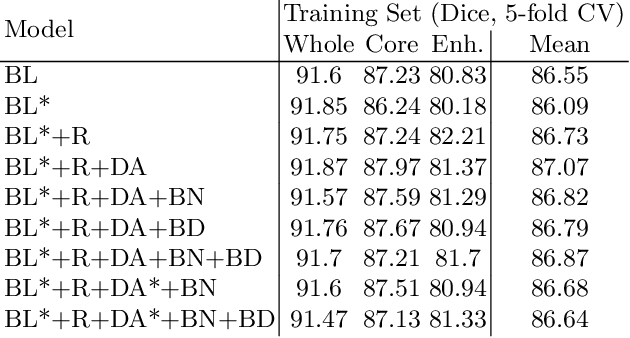
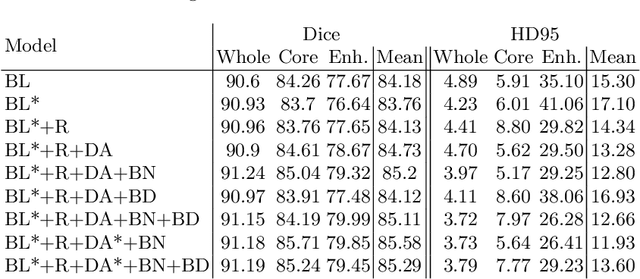
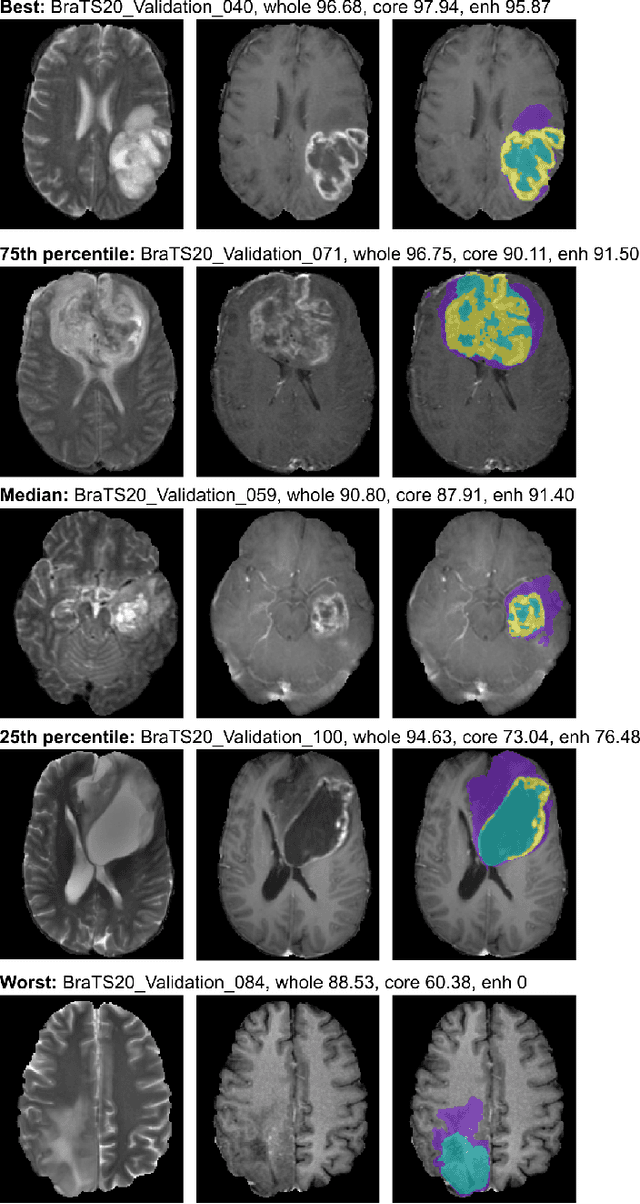
Abstract:We apply nnU-Net to the segmentation task of the BraTS 2020 challenge. The unmodified nnU-Net baseline configuration already achieves a respectable result. By incorporating BraTS-specific modifications regarding postprocessing, region-based training, a more aggressive data augmentation as well as several minor modifications to the nnUNet pipeline we are able to improve its segmentation performance substantially. We furthermore re-implement the BraTS ranking scheme to determine which of our nnU-Net variants best fits the requirements imposed by it. Our final ensemble took the first place in the BraTS 2020 competition with Dice scores of 88.95, 85.06 and 82.03 and HD95 values of 8.498,17.337 and 17.805 for whole tumor, tumor core and enhancing tumor, respectively.
Heidelberg Colorectal Data Set for Surgical Data Science in the Sensor Operating Room
May 28, 2020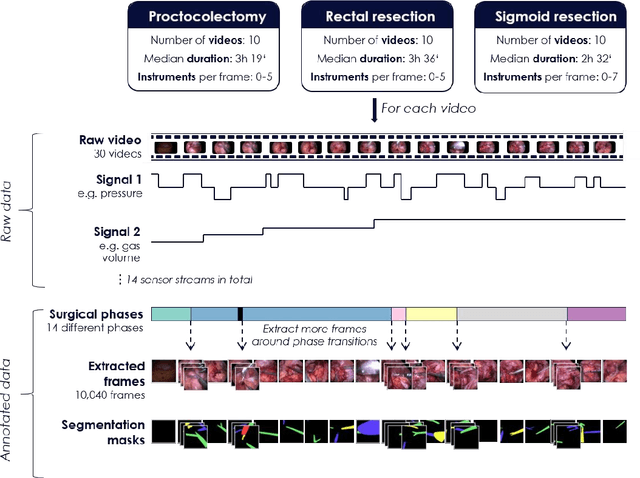
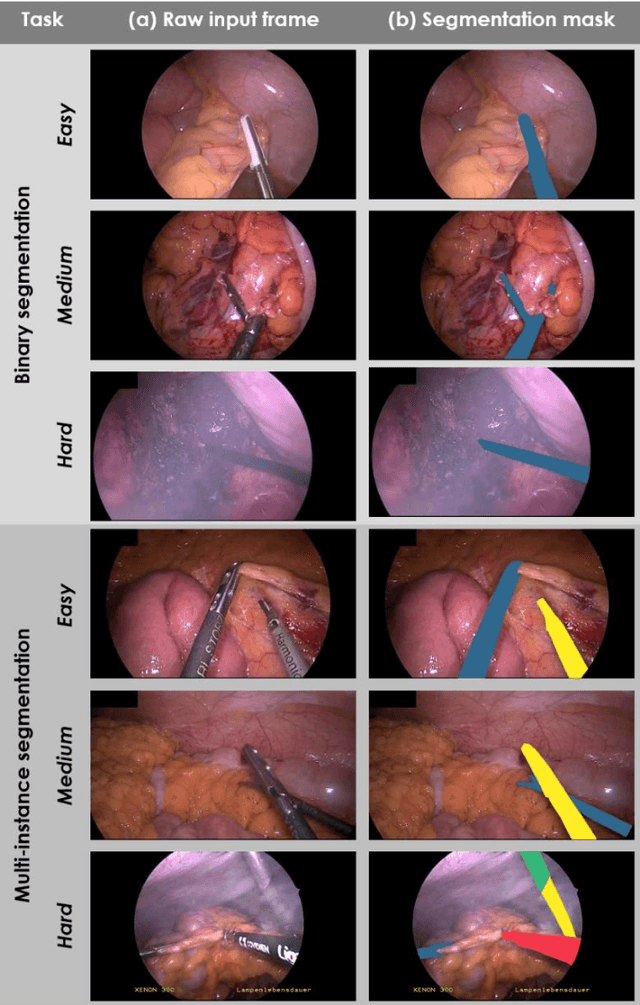
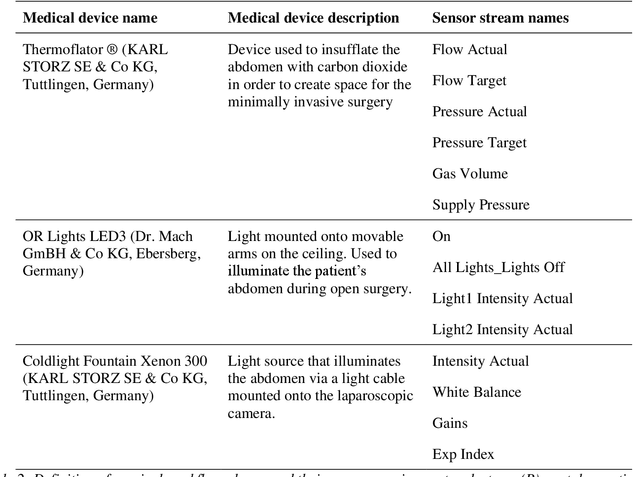
Abstract:Image-based tracking of medical instruments is an integral part of many surgical data science applications. Previous research has addressed the tasks of detecting, segmenting and tracking medical instruments based on laparoscopic video data. However, the methods proposed still tend to fail when applied to challenging images and do not generalize well to data they have not been trained on. This paper introduces the Heidelberg Colorectal (HeiCo) data set - the first publicly available data set enabling comprehensive benchmarking of medical instrument detection and segmentation algorithms with a specific emphasis on robustness and generalization capabilities of the methods. Our data set comprises 30 laparoscopic videos and corresponding sensor data from medical devices in the operating room for three different types of laparoscopic surgery. Annotations include surgical phase labels for all frames in the videos as well as instance-wise segmentation masks for surgical instruments in more than 10,000 individual frames. The data has successfully been used to organize international competitions in the scope of the Endoscopic Vision Challenges (EndoVis) 2017 and 2019.
Robust Medical Instrument Segmentation Challenge 2019
Mar 23, 2020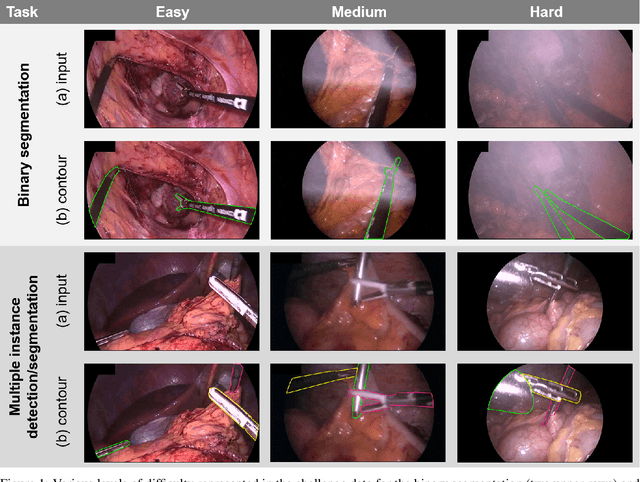
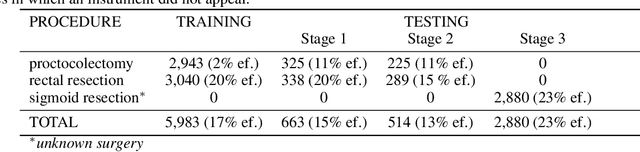
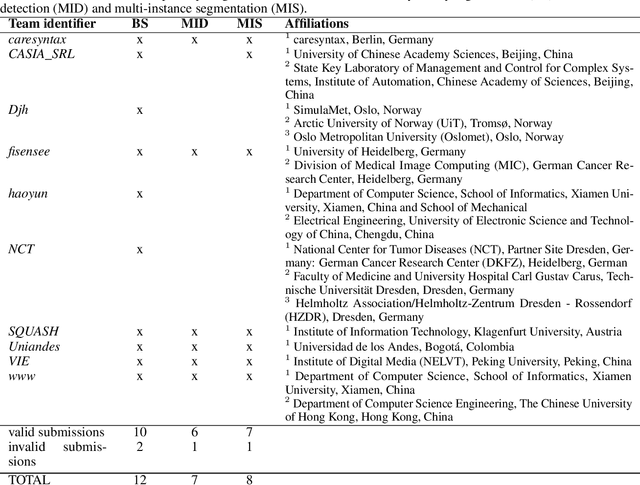
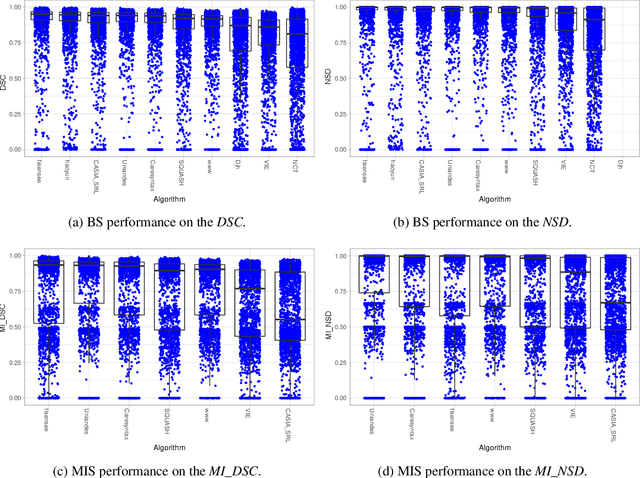
Abstract:Intraoperative tracking of laparoscopic instruments is often a prerequisite for computer and robotic-assisted interventions. While numerous methods for detecting, segmenting and tracking of medical instruments based on endoscopic video images have been proposed in the literature, key limitations remain to be addressed: Firstly, robustness, that is, the reliable performance of state-of-the-art methods when run on challenging images (e.g. in the presence of blood, smoke or motion artifacts). Secondly, generalization; algorithms trained for a specific intervention in a specific hospital should generalize to other interventions or institutions. In an effort to promote solutions for these limitations, we organized the Robust Medical Instrument Segmentation (ROBUST-MIS) challenge as an international benchmarking competition with a specific focus on the robustness and generalization capabilities of algorithms. For the first time in the field of endoscopic image processing, our challenge included a task on binary segmentation and also addressed multi-instance detection and segmentation. The challenge was based on a surgical data set comprising 10,040 annotated images acquired from a total of 30 surgical procedures from three different types of surgery. The validation of the competing methods for the three tasks (binary segmentation, multi-instance detection and multi-instance segmentation) was performed in three different stages with an increasing domain gap between the training and the test data. The results confirm the initial hypothesis, namely that algorithm performance degrades with an increasing domain gap. While the average detection and segmentation quality of the best-performing algorithms is high, future research should concentrate on detection and segmentation of small, crossing, moving and transparent instrument(s) (parts).
Is the winner really the best? A critical analysis of common research practice in biomedical image analysis competitions
Jun 06, 2018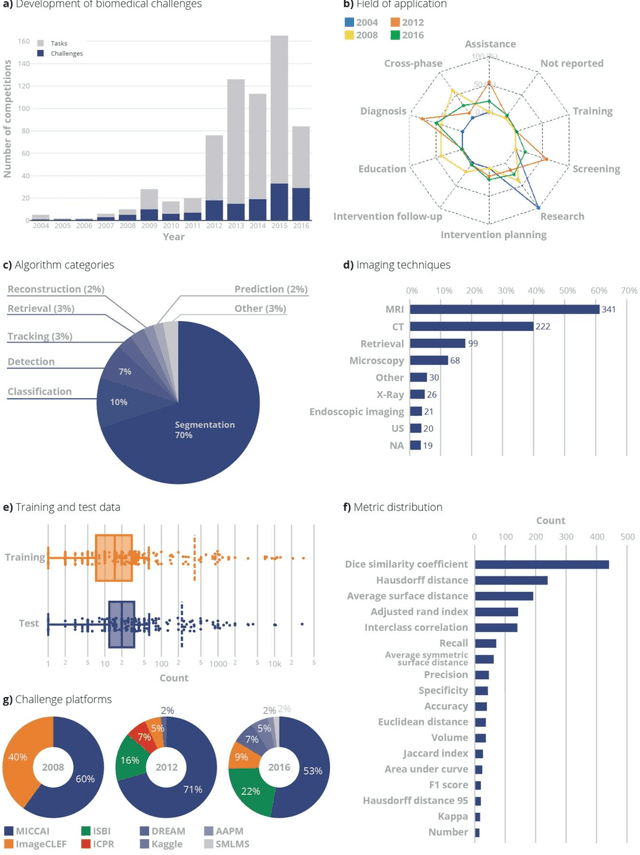
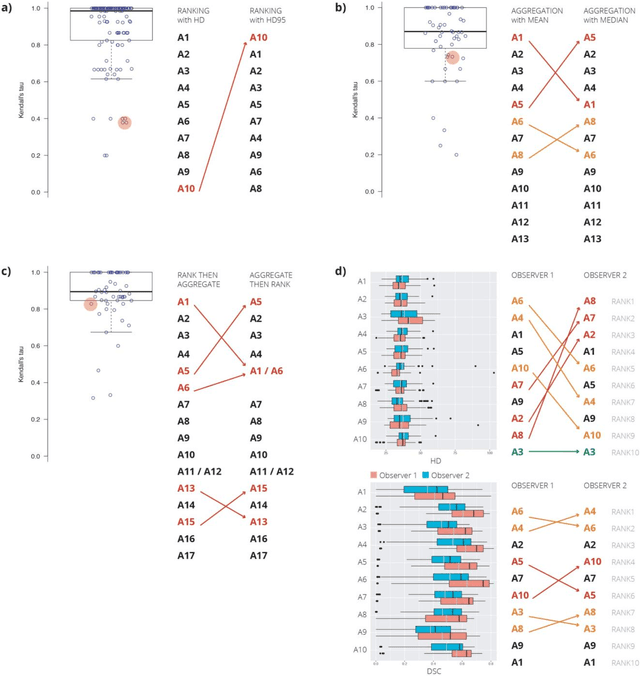
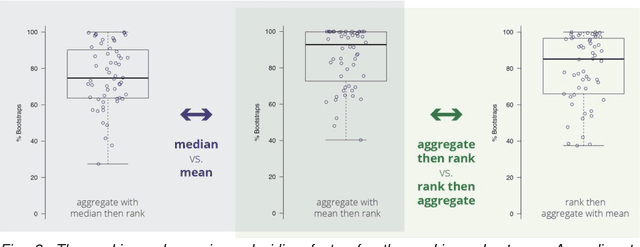
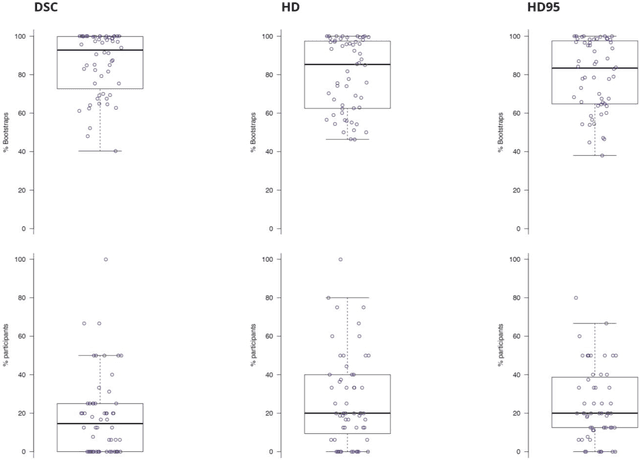
Abstract:International challenges have become the standard for validation of biomedical image analysis methods. Given their scientific impact, it is surprising that a critical analysis of common practices related to the organization of challenges has not yet been performed. In this paper, we present a comprehensive analysis of biomedical image analysis challenges conducted up to now. We demonstrate the importance of challenges and show that the lack of quality control has critical consequences. First, reproducibility and interpretation of the results is often hampered as only a fraction of relevant information is typically provided. Second, the rank of an algorithm is generally not robust to a number of variables such as the test data used for validation, the ranking scheme applied and the observers that make the reference annotations. To overcome these problems, we recommend best practice guidelines and define open research questions to be addressed in the future.
Automatic Cardiac Disease Assessment on cine-MRI via Time-Series Segmentation and Domain Specific Features
Jan 25, 2018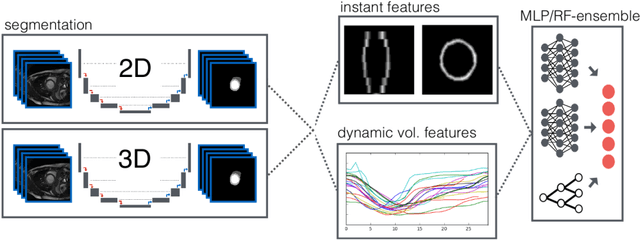
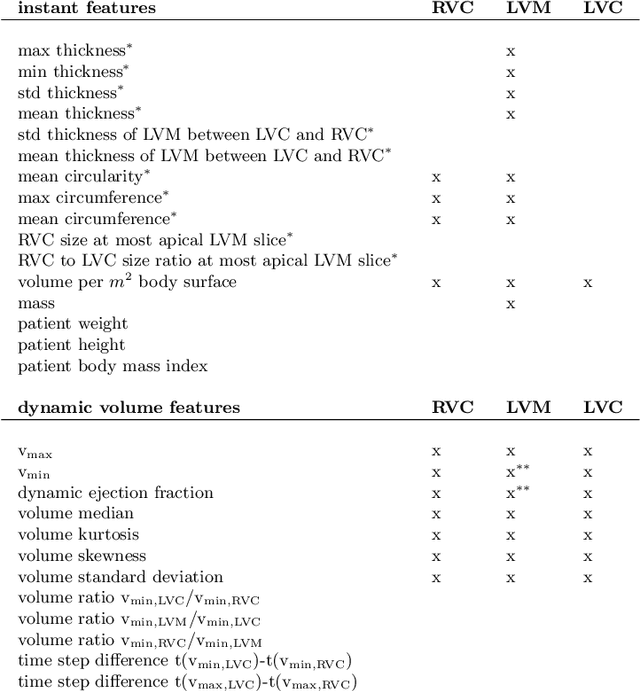
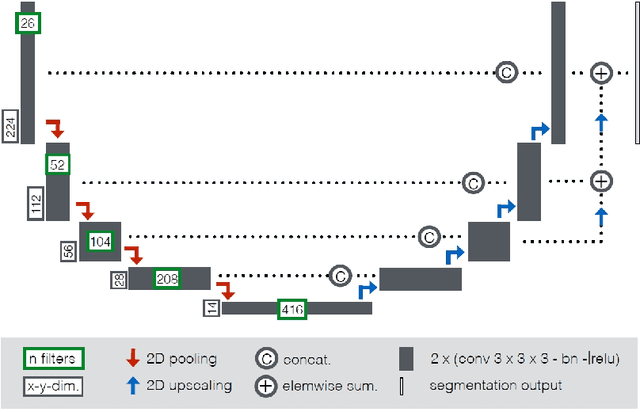
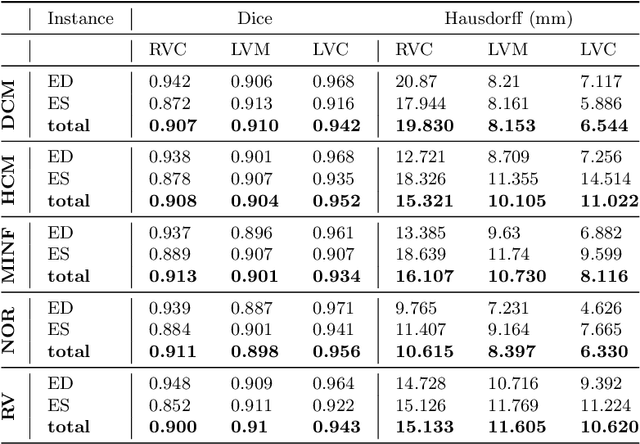
Abstract:Cardiac magnetic resonance imaging improves on diagnosis of cardiovascular diseases by providing images at high spatiotemporal resolution. Manual evaluation of these time-series, however, is expensive and prone to biased and non-reproducible outcomes. In this paper, we present a method that addresses named limitations by integrating segmentation and disease classification into a fully automatic processing pipeline. We use an ensemble of UNet inspired architectures for segmentation of cardiac structures such as the left and right ventricular cavity (LVC, RVC) and the left ventricular myocardium (LVM) on each time instance of the cardiac cycle. For the classification task, information is extracted from the segmented time-series in form of comprehensive features handcrafted to reflect diagnostic clinical procedures. Based on these features we train an ensemble of heavily regularized multilayer perceptrons (MLP) and a random forest classifier to predict the pathologic target class. We evaluated our method on the ACDC dataset (4 pathology groups, 1 healthy group) and achieve dice scores of 0.945 (LVC), 0.908 (RVC) and 0.905 (LVM) in a cross-validation over the training set (100 cases) and 0.950 (LVC), 0.923 (RVC) and 0.911 (LVM) on the test set (50 cases). We report a classification accuracy of 94% on a training set cross-validation and 92% on the test set. Our results underpin the potential of machine learning methods for accurate, fast and reproducible segmentation and computer-assisted diagnosis (CAD).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge