Klaus H. Maier-Hein
Division of Medical Image Computing, German Cancer Research Center, Heidelberg, Germany
In search of truth: Evaluating concordance of AI-based anatomy segmentation models
Dec 17, 2025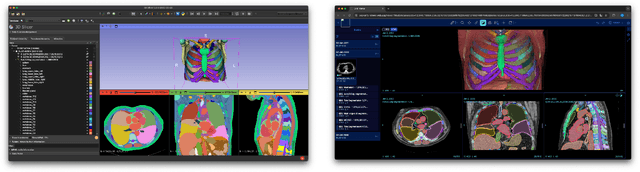


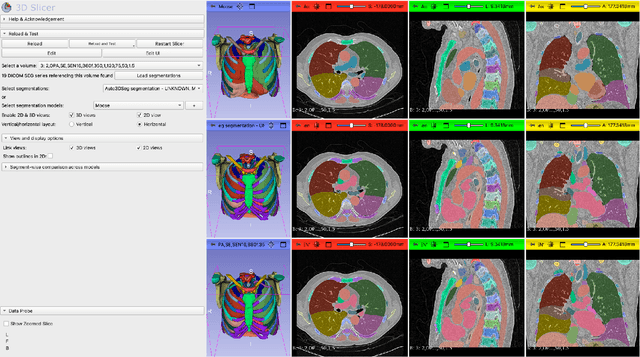
Abstract:Purpose AI-based methods for anatomy segmentation can help automate characterization of large imaging datasets. The growing number of similar in functionality models raises the challenge of evaluating them on datasets that do not contain ground truth annotations. We introduce a practical framework to assist in this task. Approach We harmonize the segmentation results into a standard, interoperable representation, which enables consistent, terminology-based labeling of the structures. We extend 3D Slicer to streamline loading and comparison of these harmonized segmentations, and demonstrate how standard representation simplifies review of the results using interactive summary plots and browser-based visualization using OHIF Viewer. To demonstrate the utility of the approach we apply it to evaluating segmentation of 31 anatomical structures (lungs, vertebrae, ribs, and heart) by six open-source models - TotalSegmentator 1.5 and 2.6, Auto3DSeg, MOOSE, MultiTalent, and CADS - for a sample of Computed Tomography (CT) scans from the publicly available National Lung Screening Trial (NLST) dataset. Results We demonstrate the utility of the framework in enabling automating loading, structure-wise inspection and comparison across models. Preliminary results ascertain practical utility of the approach in allowing quick detection and review of problematic results. The comparison shows excellent agreement segmenting some (e.g., lung) but not all structures (e.g., some models produce invalid vertebrae or rib segmentations). Conclusions The resources developed are linked from https://imagingdatacommons.github.io/segmentation-comparison/ including segmentation harmonization scripts, summary plots, and visualization tools. This work assists in model evaluation in absence of ground truth, ultimately enabling informed model selection.
The Missing Piece: A Case for Pre-Training in 3D Medical Object Detection
Sep 19, 2025Abstract:Large-scale pre-training holds the promise to advance 3D medical object detection, a crucial component of accurate computer-aided diagnosis. Yet, it remains underexplored compared to segmentation, where pre-training has already demonstrated significant benefits. Existing pre-training approaches for 3D object detection rely on 2D medical data or natural image pre-training, failing to fully leverage 3D volumetric information. In this work, we present the first systematic study of how existing pre-training methods can be integrated into state-of-the-art detection architectures, covering both CNNs and Transformers. Our results show that pre-training consistently improves detection performance across various tasks and datasets. Notably, reconstruction-based self-supervised pre-training outperforms supervised pre-training, while contrastive pre-training provides no clear benefit for 3D medical object detection. Our code is publicly available at: https://github.com/MIC-DKFZ/nnDetection-finetuning.
* MICCAI 2025
Towards Interactive Lesion Segmentation in Whole-Body PET/CT with Promptable Models
Aug 29, 2025Abstract:Whole-body PET/CT is a cornerstone of oncological imaging, yet accurate lesion segmentation remains challenging due to tracer heterogeneity, physiological uptake, and multi-center variability. While fully automated methods have advanced substantially, clinical practice benefits from approaches that keep humans in the loop to efficiently refine predicted masks. The autoPET/CT IV challenge addresses this need by introducing interactive segmentation tasks based on simulated user prompts. In this work, we present our submission to Task 1. Building on the winning autoPET III nnU-Net pipeline, we extend the framework with promptable capabilities by encoding user-provided foreground and background clicks as additional input channels. We systematically investigate representations for spatial prompts and demonstrate that Euclidean Distance Transform (EDT) encodings consistently outperform Gaussian kernels. Furthermore, we propose online simulation of user interactions and a custom point sampling strategy to improve robustness under realistic prompting conditions. Our ensemble of EDT-based models, trained with and without external data, achieves the strongest cross-validation performance, reducing both false positives and false negatives compared to baseline models. These results highlight the potential of promptable models to enable efficient, user-guided segmentation workflows in multi-tracer, multi-center PET/CT. Code is publicly available at https://github.com/MIC-DKFZ/autoPET-interactive
A Multi-Stage Fine-Tuning and Ensembling Strategy for Pancreatic Tumor Segmentation in Diagnostic and Therapeutic MRI
Aug 29, 2025Abstract:Automated segmentation of Pancreatic Ductal Adenocarcinoma (PDAC) from MRI is critical for clinical workflows but is hindered by poor tumor-tissue contrast and a scarcity of annotated data. This paper details our submission to the PANTHER challenge, addressing both diagnostic T1-weighted (Task 1) and therapeutic T2-weighted (Task 2) segmentation. Our approach is built upon the nnU-Net framework and leverages a deep, multi-stage cascaded pre-training strategy, starting from a general anatomical foundation model and sequentially fine-tuning on CT pancreatic lesion datasets and the target MRI modalities. Through extensive five-fold cross-validation, we systematically evaluated data augmentation schemes and training schedules. Our analysis revealed a critical trade-off, where aggressive data augmentation produced the highest volumetric accuracy, while default augmentations yielded superior boundary precision (achieving a state-of-the-art MASD of 5.46 mm and HD95 of 17.33 mm for Task 1). For our final submission, we exploited this finding by constructing custom, heterogeneous ensembles of specialist models, essentially creating a mix of experts. This metric-aware ensembling strategy proved highly effective, achieving a top cross-validation Tumor Dice score of 0.661 for Task 1 and 0.523 for Task 2. Our work presents a robust methodology for developing specialized, high-performance models in the context of limited data and complex medical imaging tasks (Team MIC-DKFZ).
Medical Image De-Identification Benchmark Challenge
Jul 31, 2025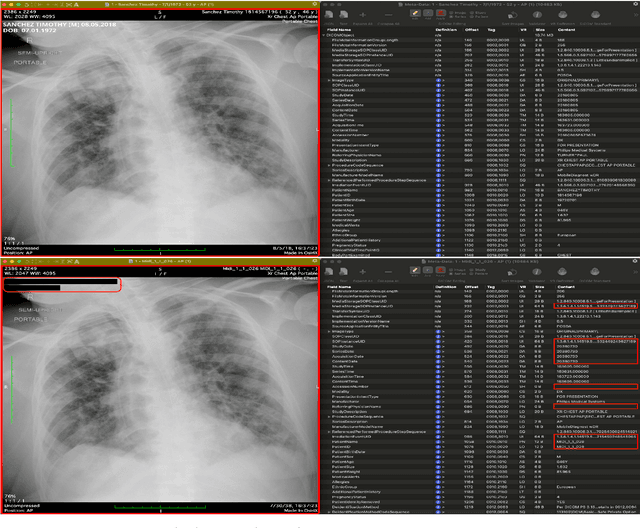
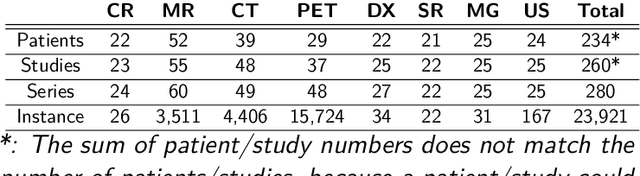
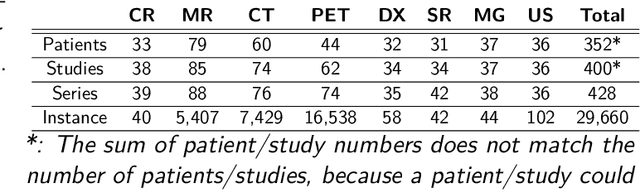
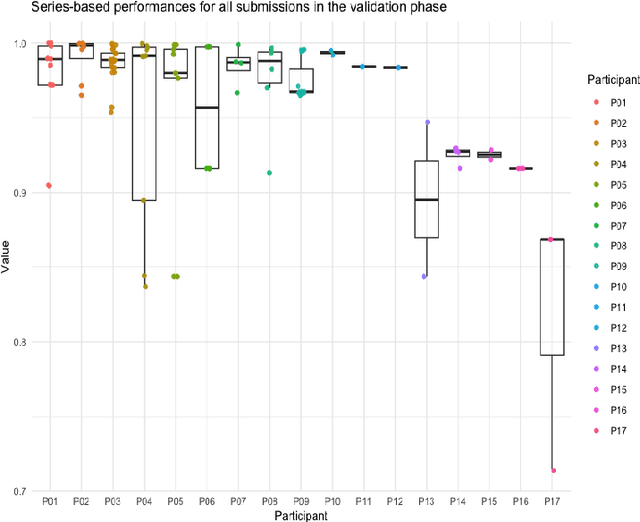
Abstract:The de-identification (deID) of protected health information (PHI) and personally identifiable information (PII) is a fundamental requirement for sharing medical images, particularly through public repositories, to ensure compliance with patient privacy laws. In addition, preservation of non-PHI metadata to inform and enable downstream development of imaging artificial intelligence (AI) is an important consideration in biomedical research. The goal of MIDI-B was to provide a standardized platform for benchmarking of DICOM image deID tools based on a set of rules conformant to the HIPAA Safe Harbor regulation, the DICOM Attribute Confidentiality Profiles, and best practices in preservation of research-critical metadata, as defined by The Cancer Imaging Archive (TCIA). The challenge employed a large, diverse, multi-center, and multi-modality set of real de-identified radiology images with synthetic PHI/PII inserted. The MIDI-B Challenge consisted of three phases: training, validation, and test. Eighty individuals registered for the challenge. In the training phase, we encouraged participants to tune their algorithms using their in-house or public data. The validation and test phases utilized the DICOM images containing synthetic identifiers (of 216 and 322 subjects, respectively). Ten teams successfully completed the test phase of the challenge. To measure success of a rule-based approach to image deID, scores were computed as the percentage of correct actions from the total number of required actions. The scores ranged from 97.91% to 99.93%. Participants employed a variety of open-source and proprietary tools with customized configurations, large language models, and optical character recognition (OCR). In this paper we provide a comprehensive report on the MIDI-B Challenge's design, implementation, results, and lessons learned.
Large Scale Supervised Pretraining For Traumatic Brain Injury Segmentation
Apr 09, 2025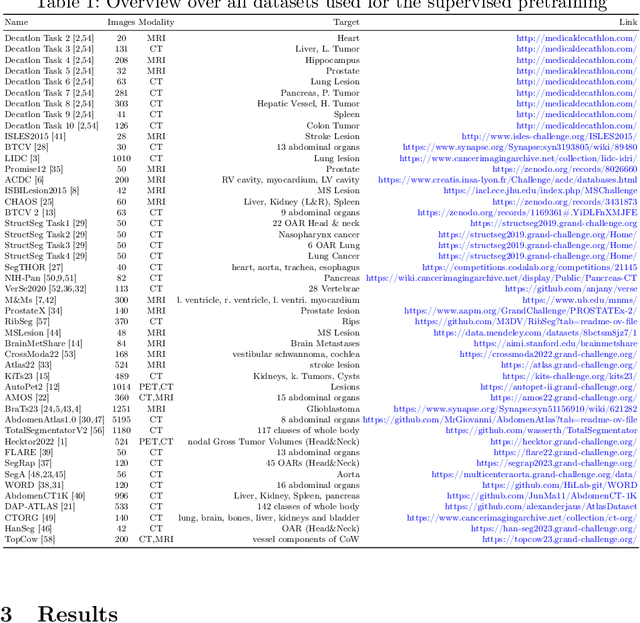
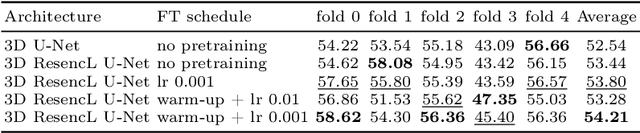
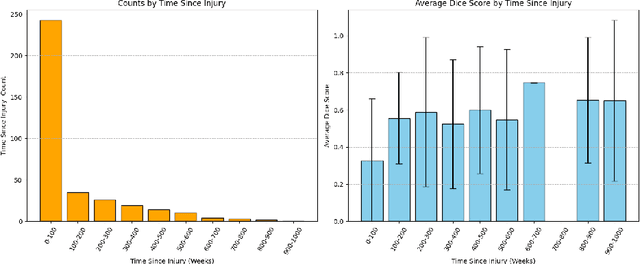
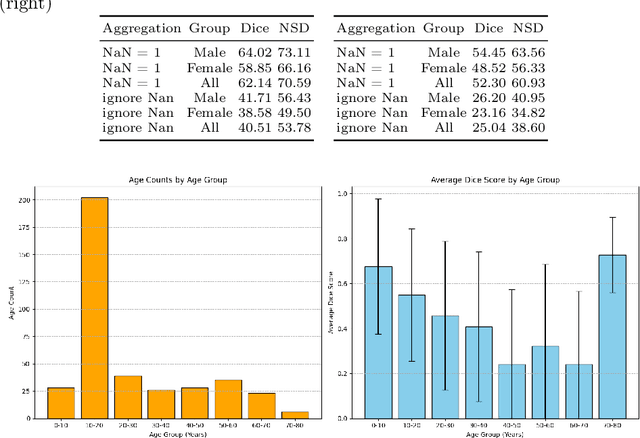
Abstract:The segmentation of lesions in Moderate to Severe Traumatic Brain Injury (msTBI) presents a significant challenge in neuroimaging due to the diverse characteristics of these lesions, which vary in size, shape, and distribution across brain regions and tissue types. This heterogeneity complicates traditional image processing techniques, resulting in critical errors in tasks such as image registration and brain parcellation. To address these challenges, the AIMS-TBI Segmentation Challenge 2024 aims to advance innovative segmentation algorithms specifically designed for T1-weighted MRI data, the most widely utilized imaging modality in clinical practice. Our proposed solution leverages a large-scale multi-dataset supervised pretraining approach inspired by the MultiTalent method. We train a Resenc L network on a comprehensive collection of datasets covering various anatomical and pathological structures, which equips the model with a robust understanding of brain anatomy and pathology. Following this, the model is fine-tuned on msTBI-specific data to optimize its performance for the unique characteristics of T1-weighted MRI scans and outperforms the baseline without pretraining up to 2 Dice points.
Multi-Class Segmentation of Aortic Branches and Zones in Computed Tomography Angiography: The AortaSeg24 Challenge
Feb 07, 2025



Abstract:Multi-class segmentation of the aorta in computed tomography angiography (CTA) scans is essential for diagnosing and planning complex endovascular treatments for patients with aortic dissections. However, existing methods reduce aortic segmentation to a binary problem, limiting their ability to measure diameters across different branches and zones. Furthermore, no open-source dataset is currently available to support the development of multi-class aortic segmentation methods. To address this gap, we organized the AortaSeg24 MICCAI Challenge, introducing the first dataset of 100 CTA volumes annotated for 23 clinically relevant aortic branches and zones. This dataset was designed to facilitate both model development and validation. The challenge attracted 121 teams worldwide, with participants leveraging state-of-the-art frameworks such as nnU-Net and exploring novel techniques, including cascaded models, data augmentation strategies, and custom loss functions. We evaluated the submitted algorithms using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD), highlighting the approaches adopted by the top five performing teams. This paper presents the challenge design, dataset details, evaluation metrics, and an in-depth analysis of the top-performing algorithms. The annotated dataset, evaluation code, and implementations of the leading methods are publicly available to support further research. All resources can be accessed at https://aortaseg24.grand-challenge.org.
An OpenMind for 3D medical vision self-supervised learning
Dec 22, 2024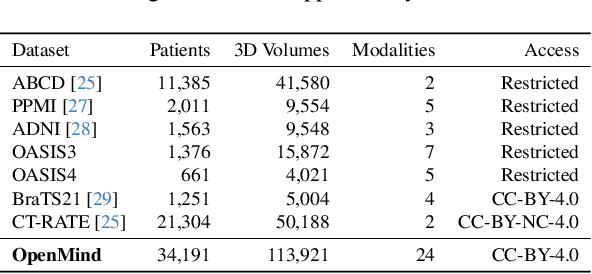
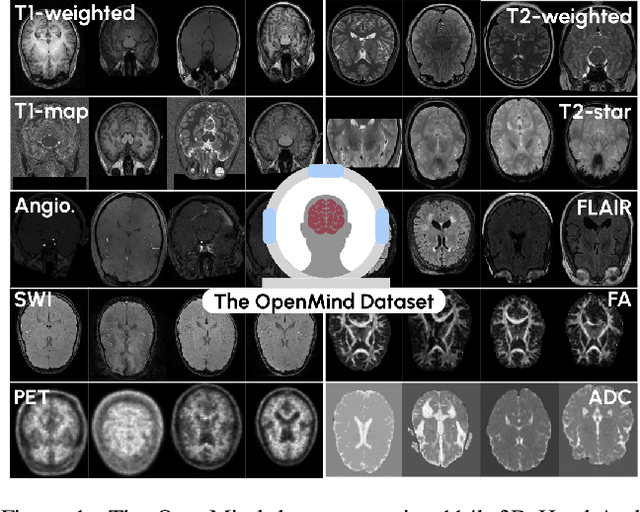
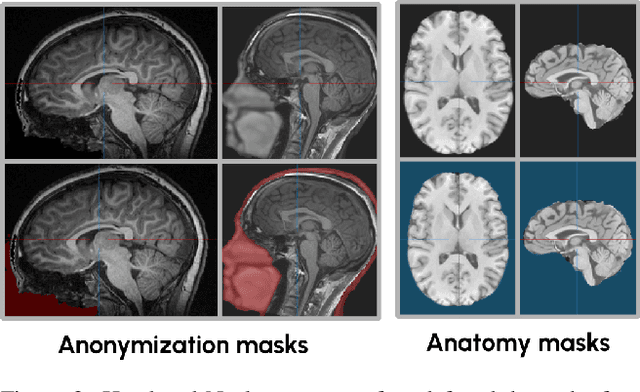
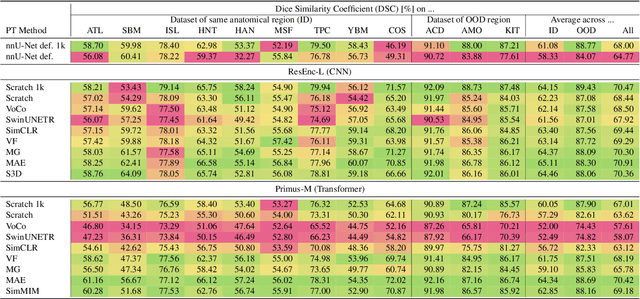
Abstract:The field of 3D medical vision self-supervised learning lacks consistency and standardization. While many methods have been developed it is impossible to identify the current state-of-the-art, due to i) varying and small pre-training datasets, ii) varying architectures, and iii) being evaluated on differing downstream datasets. In this paper we bring clarity to this field and lay the foundation for further method advancements: We a) publish the largest publicly available pre-training dataset comprising 114k 3D brain MRI volumes and b) benchmark existing SSL methods under common architectures and c) provide the code of our framework publicly to facilitate rapid adoption and reproduction. This pre-print \textit{only describes} the dataset contribution (a); Data, benchmark, and codebase will be made available shortly.
Scaling nnU-Net for CBCT Segmentation
Nov 26, 2024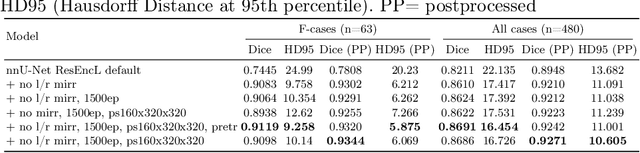
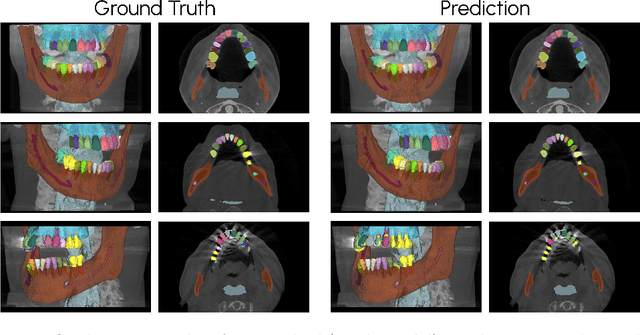
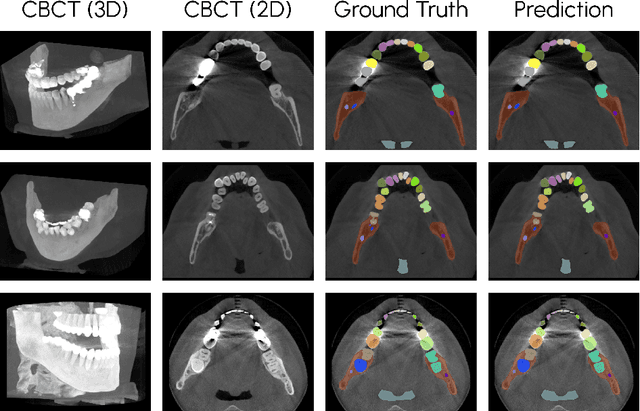
Abstract:This paper presents our approach to scaling the nnU-Net framework for multi-structure segmentation on Cone Beam Computed Tomography (CBCT) images, specifically in the scope of the ToothFairy2 Challenge. We leveraged the nnU-Net ResEnc L model, introducing key modifications to patch size, network topology, and data augmentation strategies to address the unique challenges of dental CBCT imaging. Our method achieved a mean Dice coefficient of 0.9253 and HD95 of 18.472 on the test set, securing a mean rank of 4.6 and with it the first place in the ToothFairy2 challenge. The source code is publicly available, encouraging further research and development in the field.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge