Paul F. Jaeger
TxGemma: Efficient and Agentic LLMs for Therapeutics
Apr 08, 2025Abstract:Therapeutic development is a costly and high-risk endeavor that is often plagued by high failure rates. To address this, we introduce TxGemma, a suite of efficient, generalist large language models (LLMs) capable of therapeutic property prediction as well as interactive reasoning and explainability. Unlike task-specific models, TxGemma synthesizes information from diverse sources, enabling broad application across the therapeutic development pipeline. The suite includes 2B, 9B, and 27B parameter models, fine-tuned from Gemma-2 on a comprehensive dataset of small molecules, proteins, nucleic acids, diseases, and cell lines. Across 66 therapeutic development tasks, TxGemma achieved superior or comparable performance to the state-of-the-art generalist model on 64 (superior on 45), and against state-of-the-art specialist models on 50 (superior on 26). Fine-tuning TxGemma models on therapeutic downstream tasks, such as clinical trial adverse event prediction, requires less training data than fine-tuning base LLMs, making TxGemma suitable for data-limited applications. Beyond these predictive capabilities, TxGemma features conversational models that bridge the gap between general LLMs and specialized property predictors. These allow scientists to interact in natural language, provide mechanistic reasoning for predictions based on molecular structure, and engage in scientific discussions. Building on this, we further introduce Agentic-Tx, a generalist therapeutic agentic system powered by Gemini 2.5 that reasons, acts, manages diverse workflows, and acquires external domain knowledge. Agentic-Tx surpasses prior leading models on the Humanity's Last Exam benchmark (Chemistry & Biology) with 52.3% relative improvement over o3-mini (high) and 26.7% over o3-mini (high) on GPQA (Chemistry) and excels with improvements of 6.3% (ChemBench-Preference) and 2.4% (ChemBench-Mini) over o3-mini (high).
Bridging vision language model (VLM) evaluation gaps with a framework for scalable and cost-effective benchmark generation
Feb 21, 2025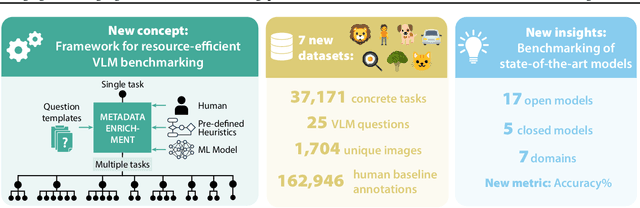
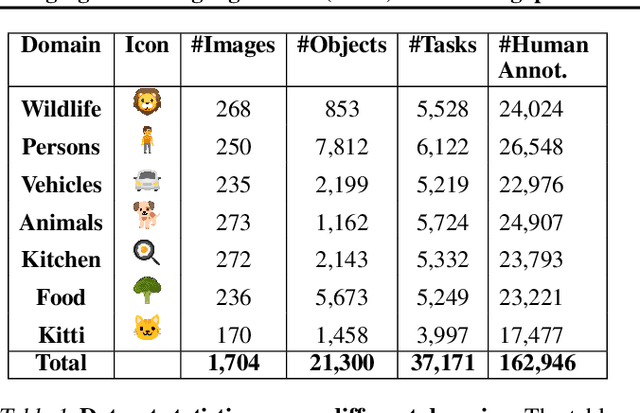
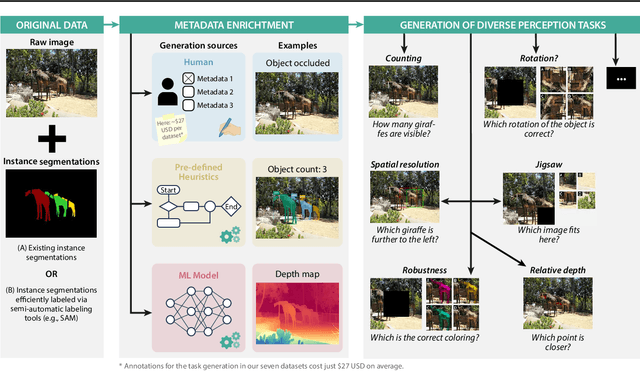
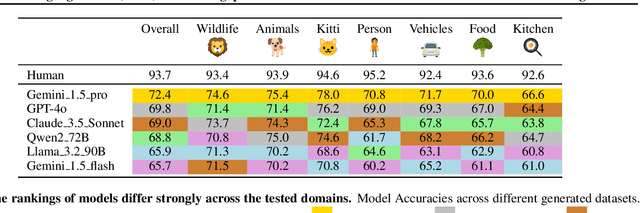
Abstract:Reliable evaluation of AI models is critical for scientific progress and practical application. While existing VLM benchmarks provide general insights into model capabilities, their heterogeneous designs and limited focus on a few imaging domains pose significant challenges for both cross-domain performance comparison and targeted domain-specific evaluation. To address this, we propose three key contributions: (1) a framework for the resource-efficient creation of domain-specific VLM benchmarks enabled by task augmentation for creating multiple diverse tasks from a single existing task, (2) the release of new VLM benchmarks for seven domains, created according to the same homogeneous protocol and including 162,946 thoroughly human-validated answers, and (3) an extensive benchmarking of 22 state-of-the-art VLMs on a total of 37,171 tasks, revealing performance variances across domains and tasks, thereby supporting the need for tailored VLM benchmarks. Adoption of our methodology will pave the way for the resource-efficient domain-specific selection of models and guide future research efforts toward addressing core open questions.
SURE-VQA: Systematic Understanding of Robustness Evaluation in Medical VQA Tasks
Nov 29, 2024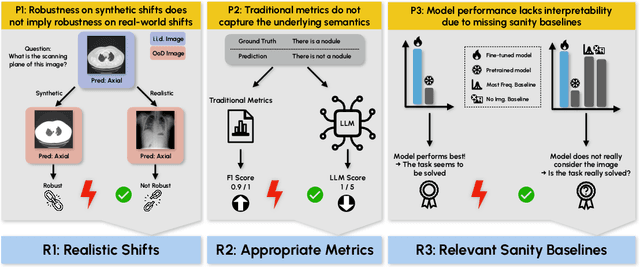
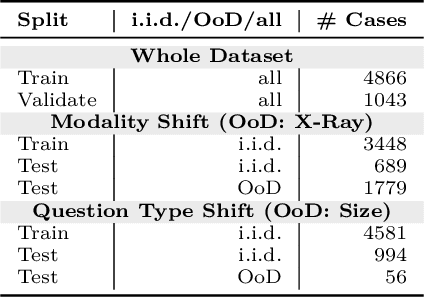
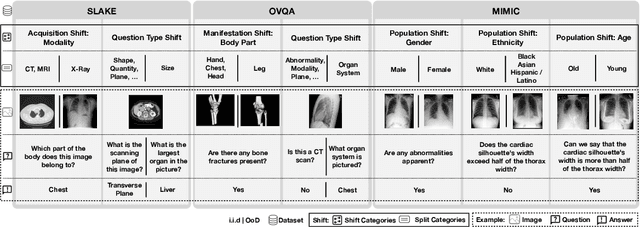
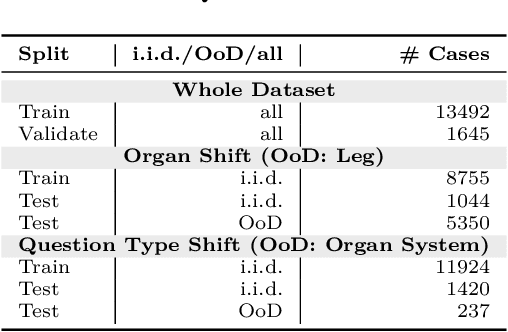
Abstract:Vision-Language Models (VLMs) have great potential in medical tasks, like Visual Question Answering (VQA), where they could act as interactive assistants for both patients and clinicians. Yet their robustness to distribution shifts on unseen data remains a critical concern for safe deployment. Evaluating such robustness requires a controlled experimental setup that allows for systematic insights into the model's behavior. However, we demonstrate that current setups fail to offer sufficiently thorough evaluations, limiting their ability to accurately assess model robustness. To address this gap, our work introduces a novel framework, called SURE-VQA, centered around three key requirements to overcome the current pitfalls and systematically analyze the robustness of VLMs: 1) Since robustness on synthetic shifts does not necessarily translate to real-world shifts, robustness should be measured on real-world shifts that are inherent to the VQA data; 2) Traditional token-matching metrics often fail to capture underlying semantics, necessitating the use of large language models (LLMs) for more accurate semantic evaluation; 3) Model performance often lacks interpretability due to missing sanity baselines, thus meaningful baselines should be reported that allow assessing the multimodal impact on the VLM. To demonstrate the relevance of this framework, we conduct a study on the robustness of various fine-tuning methods across three medical datasets with four different types of distribution shifts. Our study reveals several important findings: 1) Sanity baselines that do not utilize image data can perform surprisingly well; 2) We confirm LoRA as the best-performing PEFT method; 3) No PEFT method consistently outperforms others in terms of robustness to shifts. Code is provided at https://github.com/IML-DKFZ/sure-vqa.
INTRABENCH: Interactive Radiological Benchmark
Nov 12, 2024



Abstract:Current interactive segmentation approaches, inspired by the success of META's Segment Anything model, have achieved notable advancements, however, they come with substantial limitations that hinder their practical application in real clinical scenarios. These include unrealistic human interaction requirements, such as slice-by-slice operations for 2D models on 3D data, a lack of iterative refinement, and insufficient evaluation experiments. These shortcomings prevent accurate assessment of model performance and lead to inconsistent outcomes across studies. IntRaBench overcomes these challenges by offering a comprehensive and reproducible framework for evaluating interactive segmentation methods in realistic, clinically relevant scenarios. It includes diverse datasets, target structures, and segmentation models, and provides a flexible codebase that allows seamless integration of new models and prompting strategies. Additionally, we introduce advanced techniques to minimize clinician interaction, ensuring fair comparisons between 2D and 3D models. By open-sourcing IntRaBench, we invite the research community to integrate their models and prompting techniques, ensuring continuous and transparent evaluation of interactive segmentation models in 3D medical imaging.
Enhancing predictive imaging biomarker discovery through treatment effect analysis
Jun 04, 2024Abstract:Identifying predictive biomarkers, which forecast individual treatment effectiveness, is crucial for personalized medicine and informs decision-making across diverse disciplines. These biomarkers are extracted from pre-treatment data, often within randomized controlled trials, and have to be distinguished from prognostic biomarkers, which are independent of treatment assignment. Our study focuses on the discovery of predictive imaging biomarkers, aiming to leverage pre-treatment images to unveil new causal relationships. Previous approaches relied on labor-intensive handcrafted or manually derived features, which may introduce biases. In response, we present a new task of discovering predictive imaging biomarkers directly from the pre-treatment images to learn relevant image features. We propose an evaluation protocol for this task to assess a model's ability to identify predictive imaging biomarkers and differentiate them from prognostic ones. It employs statistical testing and a comprehensive analysis of image feature attribution. We explore the suitability of deep learning models originally designed for estimating the conditional average treatment effect (CATE) for this task, which previously have been primarily assessed for the precision of CATE estimation, overlooking the evaluation of imaging biomarker discovery. Our proof-of-concept analysis demonstrates promising results in discovering and validating predictive imaging biomarkers from synthetic outcomes and real-world image datasets.
nnU-Net Revisited: A Call for Rigorous Validation in 3D Medical Image Segmentation
Apr 15, 2024



Abstract:The release of nnU-Net marked a paradigm shift in 3D medical image segmentation, demonstrating that a properly configured U-Net architecture could still achieve state-of-the-art results. Despite this, the pursuit of novel architectures, and the respective claims of superior performance over the U-Net baseline, continued. In this study, we demonstrate that many of these recent claims fail to hold up when scrutinized for common validation shortcomings, such as the use of inadequate baselines, insufficient datasets, and neglected computational resources. By meticulously avoiding these pitfalls, we conduct a thorough and comprehensive benchmarking of current segmentation methods including CNN-based, Transformer-based, and Mamba-based approaches. In contrast to current beliefs, we find that the recipe for state-of-the-art performance is 1) employing CNN-based U-Net models, including ResNet and ConvNeXt variants, 2) using the nnU-Net framework, and 3) scaling models to modern hardware resources. These results indicate an ongoing innovation bias towards novel architectures in the field and underscore the need for more stringent validation standards in the quest for scientific progress.
ValUES: A Framework for Systematic Validation of Uncertainty Estimation in Semantic Segmentation
Jan 16, 2024



Abstract:Uncertainty estimation is an essential and heavily-studied component for the reliable application of semantic segmentation methods. While various studies exist claiming methodological advances on the one hand, and successful application on the other hand, the field is currently hampered by a gap between theory and practice leaving fundamental questions unanswered: Can data-related and model-related uncertainty really be separated in practice? Which components of an uncertainty method are essential for real-world performance? Which uncertainty method works well for which application? In this work, we link this research gap to a lack of systematic and comprehensive evaluation of uncertainty methods. Specifically, we identify three key pitfalls in current literature and present an evaluation framework that bridges the research gap by providing 1) a controlled environment for studying data ambiguities as well as distribution shifts, 2) systematic ablations of relevant method components, and 3) test-beds for the five predominant uncertainty applications: OoD-detection, active learning, failure detection, calibration, and ambiguity modeling. Empirical results on simulated as well as real-world data demonstrate how the proposed framework is able to answer the predominant questions in the field revealing for instance that 1) separation of uncertainty types works on simulated data but does not necessarily translate to real-world data, 2) aggregation of scores is a crucial but currently neglected component of uncertainty methods, 3) While ensembles are performing most robustly across the different downstream tasks and settings, test-time augmentation often constitutes a light-weight alternative. Code is at: https://github.com/IML-DKFZ/values
Application-driven Validation of Posteriors in Inverse Problems
Sep 18, 2023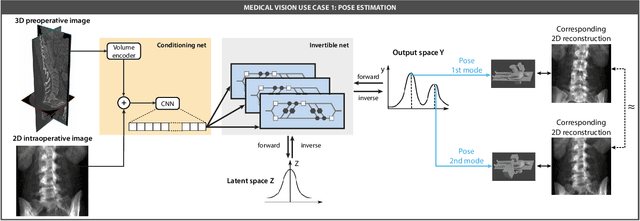
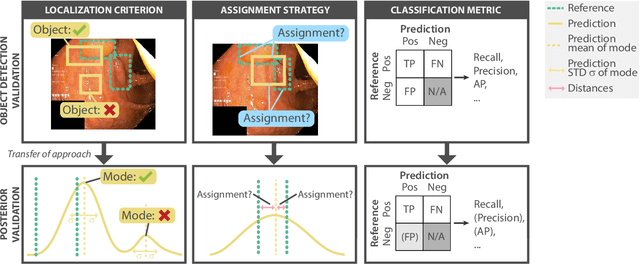
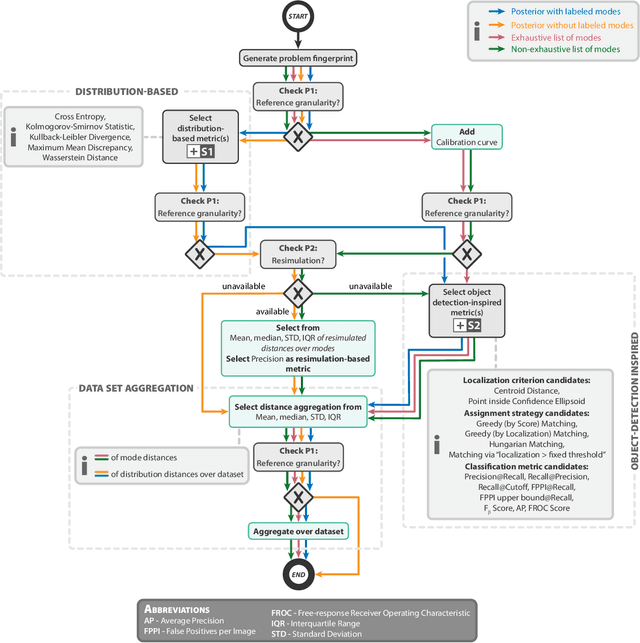
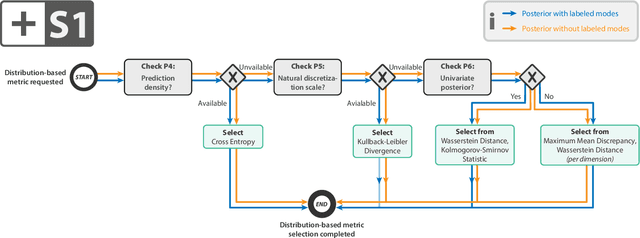
Abstract:Current deep learning-based solutions for image analysis tasks are commonly incapable of handling problems to which multiple different plausible solutions exist. In response, posterior-based methods such as conditional Diffusion Models and Invertible Neural Networks have emerged; however, their translation is hampered by a lack of research on adequate validation. In other words, the way progress is measured often does not reflect the needs of the driving practical application. Closing this gap in the literature, we present the first systematic framework for the application-driven validation of posterior-based methods in inverse problems. As a methodological novelty, it adopts key principles from the field of object detection validation, which has a long history of addressing the question of how to locate and match multiple object instances in an image. Treating modes as instances enables us to perform mode-centric validation, using well-interpretable metrics from the application perspective. We demonstrate the value of our framework through instantiations for a synthetic toy example and two medical vision use cases: pose estimation in surgery and imaging-based quantification of functional tissue parameters for diagnostics. Our framework offers key advantages over common approaches to posterior validation in all three examples and could thus revolutionize performance assessment in inverse problems.
RecycleNet: Latent Feature Recycling Leads to Iterative Decision Refinement
Sep 14, 2023



Abstract:Despite the remarkable success of deep learning systems over the last decade, a key difference still remains between neural network and human decision-making: As humans, we cannot only form a decision on the spot, but also ponder, revisiting an initial guess from different angles, distilling relevant information, arriving at a better decision. Here, we propose RecycleNet, a latent feature recycling method, instilling the pondering capability for neural networks to refine initial decisions over a number of recycling steps, where outputs are fed back into earlier network layers in an iterative fashion. This approach makes minimal assumptions about the neural network architecture and thus can be implemented in a wide variety of contexts. Using medical image segmentation as the evaluation environment, we show that latent feature recycling enables the network to iteratively refine initial predictions even beyond the iterations seen during training, converging towards an improved decision. We evaluate this across a variety of segmentation benchmarks and show consistent improvements even compared with top-performing segmentation methods. This allows trading increased computation time for improved performance, which can be beneficial, especially for safety-critical applications.
Anatomy-informed Data Augmentation for Enhanced Prostate Cancer Detection
Sep 07, 2023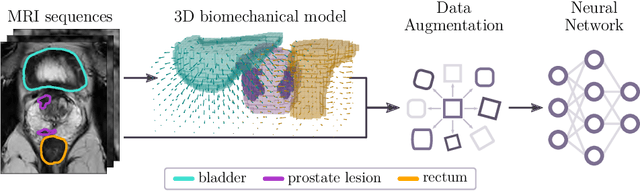

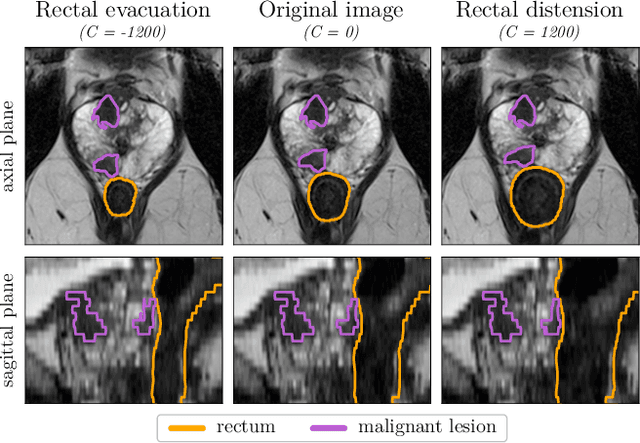
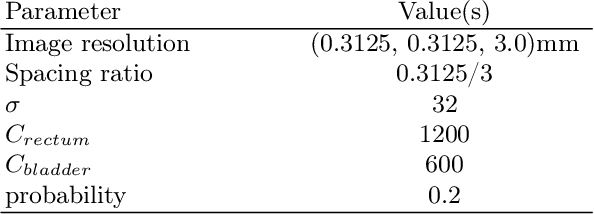
Abstract:Data augmentation (DA) is a key factor in medical image analysis, such as in prostate cancer (PCa) detection on magnetic resonance images. State-of-the-art computer-aided diagnosis systems still rely on simplistic spatial transformations to preserve the pathological label post transformation. However, such augmentations do not substantially increase the organ as well as tumor shape variability in the training set, limiting the model's ability to generalize to unseen cases with more diverse localized soft-tissue deformations. We propose a new anatomy-informed transformation that leverages information from adjacent organs to simulate typical physiological deformations of the prostate and generates unique lesion shapes without altering their label. Due to its lightweight computational requirements, it can be easily integrated into common DA frameworks. We demonstrate the effectiveness of our augmentation on a dataset of 774 biopsy-confirmed examinations, by evaluating a state-of-the-art method for PCa detection with different augmentation settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge