Annika Reinke
German Cancer Research Center, DKFZ Heidelberg, Helmholtz Imaging, Germany
Performance uncertainty in medical image analysis: a large-scale investigation of confidence intervals
Jan 23, 2026Abstract:Performance uncertainty quantification is essential for reliable validation and eventual clinical translation of medical imaging artificial intelligence (AI). Confidence intervals (CIs) play a central role in this process by indicating how precise a reported performance estimate is. Yet, due to the limited amount of work examining CI behavior in medical imaging, the community remains largely unaware of how many diverse CI methods exist and how they behave in specific settings. The purpose of this study is to close this gap. To this end, we conducted a large-scale empirical analysis across a total of 24 segmentation and classification tasks, using 19 trained models per task group, a broad spectrum of commonly used performance metrics, multiple aggregation strategies, and several widely adopted CI methods. Reliability (coverage) and precision (width) of each CI method were estimated across all settings to characterize their dependence on study characteristics. Our analysis revealed five principal findings: 1) the sample size required for reliable CIs varies from a few dozens to several thousands of cases depending on study parameters; 2) CI behavior is strongly affected by the choice of performance metric; 3) aggregation strategy substantially influences the reliability of CIs, e.g. they require more observations for macro than for micro; 4) the machine learning problem (segmentation versus classification) modulates these effects; 5) different CI methods are not equally reliable and precise depending on the use case. These results form key components for the development of future guidelines on reporting performance uncertainty in medical imaging AI.
Medical Imaging AI Competitions Lack Fairness
Dec 19, 2025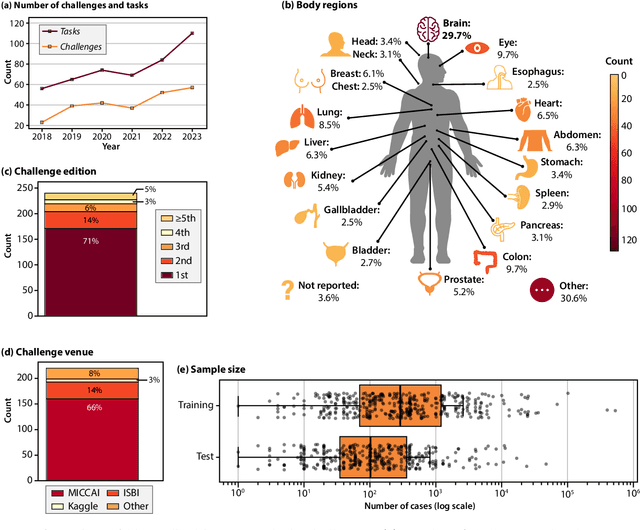
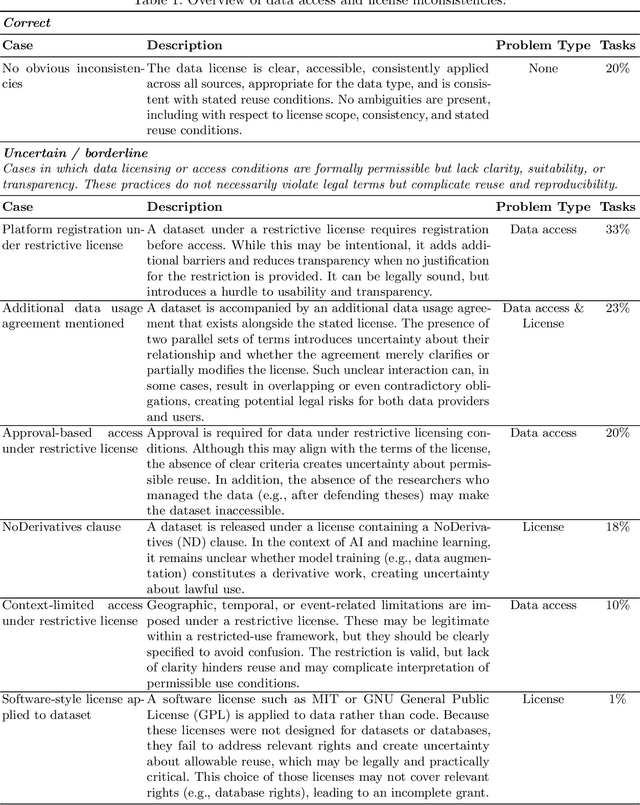
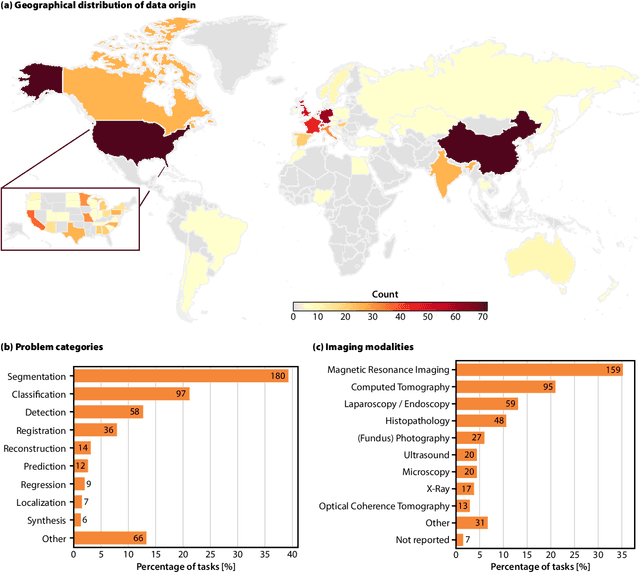
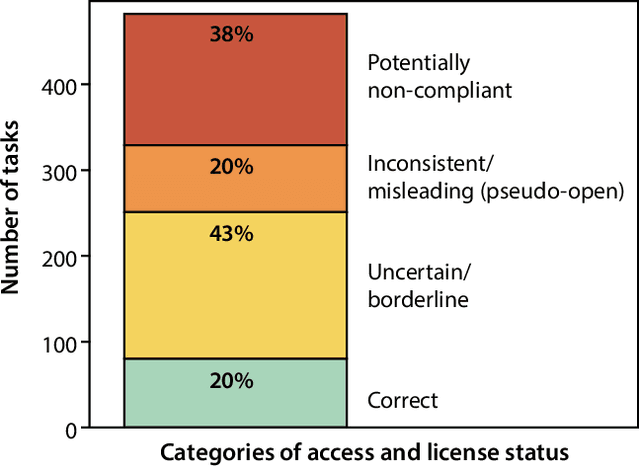
Abstract:Benchmarking competitions are central to the development of artificial intelligence (AI) in medical imaging, defining performance standards and shaping methodological progress. However, it remains unclear whether these benchmarks provide data that are sufficiently representative, accessible, and reusable to support clinically meaningful AI. In this work, we assess fairness along two complementary dimensions: (1) whether challenge datasets are representative of real-world clinical diversity, and (2) whether they are accessible and legally reusable in line with the FAIR principles. To address this question, we conducted a large-scale systematic study of 241 biomedical image analysis challenges comprising 458 tasks across 19 imaging modalities. Our findings show substantial biases in dataset composition, including geographic location, modality-, and problem type-related biases, indicating that current benchmarks do not adequately reflect real-world clinical diversity. Despite their widespread influence, challenge datasets were frequently constrained by restrictive or ambiguous access conditions, inconsistent or non-compliant licensing practices, and incomplete documentation, limiting reproducibility and long-term reuse. Together, these shortcomings expose foundational fairness limitations in our benchmarking ecosystem and highlight a disconnect between leaderboard success and clinical relevance.
Federated Learning for Surgical Vision in Appendicitis Classification: Results of the FedSurg EndoVis 2024 Challenge
Oct 06, 2025Abstract:Purpose: The FedSurg challenge was designed to benchmark the state of the art in federated learning for surgical video classification. Its goal was to assess how well current methods generalize to unseen clinical centers and adapt through local fine-tuning while enabling collaborative model development without sharing patient data. Methods: Participants developed strategies to classify inflammation stages in appendicitis using a preliminary version of the multi-center Appendix300 video dataset. The challenge evaluated two tasks: generalization to an unseen center and center-specific adaptation after fine-tuning. Submitted approaches included foundation models with linear probing, metric learning with triplet loss, and various FL aggregation schemes (FedAvg, FedMedian, FedSAM). Performance was assessed using F1-score and Expected Cost, with ranking robustness evaluated via bootstrapping and statistical testing. Results: In the generalization task, performance across centers was limited. In the adaptation task, all teams improved after fine-tuning, though ranking stability was low. The ViViT-based submission achieved the strongest overall performance. The challenge highlighted limitations in generalization, sensitivity to class imbalance, and difficulties in hyperparameter tuning in decentralized training, while spatiotemporal modeling and context-aware preprocessing emerged as promising strategies. Conclusion: The FedSurg Challenge establishes the first benchmark for evaluating FL strategies in surgical video classification. Findings highlight the trade-off between local personalization and global robustness, and underscore the importance of architecture choice, preprocessing, and loss design. This benchmarking offers a reference point for future development of imbalance-aware, adaptive, and robust FL methods in clinical surgical AI.
Challenging Vision-Language Models with Surgical Data: A New Dataset and Broad Benchmarking Study
Jun 06, 2025Abstract:While traditional computer vision models have historically struggled to generalize to endoscopic domains, the emergence of foundation models has shown promising cross-domain performance. In this work, we present the first large-scale study assessing the capabilities of Vision Language Models (VLMs) for endoscopic tasks with a specific focus on laparoscopic surgery. Using a diverse set of state-of-the-art models, multiple surgical datasets, and extensive human reference annotations, we address three key research questions: (1) Can current VLMs solve basic perception tasks on surgical images? (2) Can they handle advanced frame-based endoscopic scene understanding tasks? and (3) How do specialized medical VLMs compare to generalist models in this context? Our results reveal that VLMs can effectively perform basic surgical perception tasks, such as object counting and localization, with performance levels comparable to general domain tasks. However, their performance deteriorates significantly when the tasks require medical knowledge. Notably, we find that specialized medical VLMs currently underperform compared to generalist models across both basic and advanced surgical tasks, suggesting that they are not yet optimized for the complexity of surgical environments. These findings highlight the need for further advancements to enable VLMs to handle the unique challenges posed by surgery. Overall, our work provides important insights for the development of next-generation endoscopic AI systems and identifies key areas for improvement in medical visual language models.
False Promises in Medical Imaging AI? Assessing Validity of Outperformance Claims
May 07, 2025Abstract:Performance comparisons are fundamental in medical imaging Artificial Intelligence (AI) research, often driving claims of superiority based on relative improvements in common performance metrics. However, such claims frequently rely solely on empirical mean performance. In this paper, we investigate whether newly proposed methods genuinely outperform the state of the art by analyzing a representative cohort of medical imaging papers. We quantify the probability of false claims based on a Bayesian approach that leverages reported results alongside empirically estimated model congruence to estimate whether the relative ranking of methods is likely to have occurred by chance. According to our results, the majority (>80%) of papers claims outperformance when introducing a new method. Our analysis further revealed a high probability (>5%) of false outperformance claims in 86% of classification papers and 53% of segmentation papers. These findings highlight a critical flaw in current benchmarking practices: claims of outperformance in medical imaging AI are frequently unsubstantiated, posing a risk of misdirecting future research efforts.
Bridging vision language model (VLM) evaluation gaps with a framework for scalable and cost-effective benchmark generation
Feb 21, 2025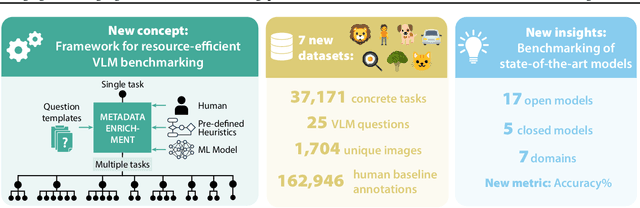
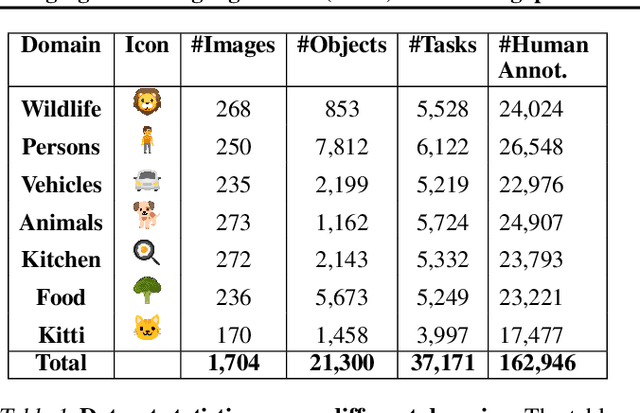
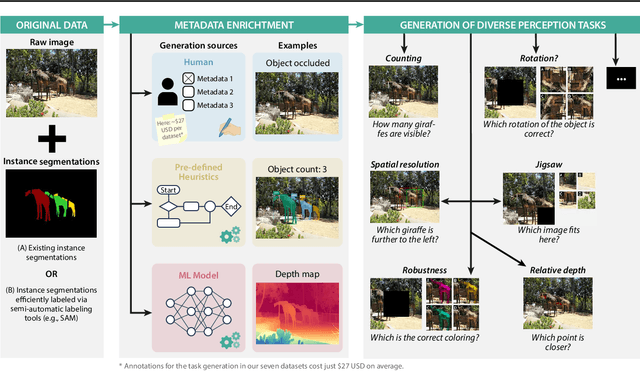
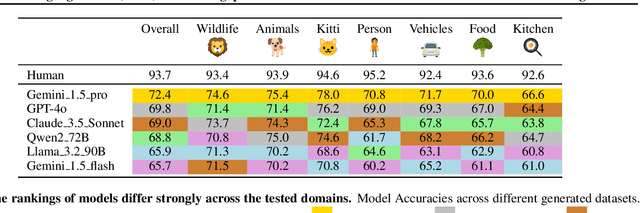
Abstract:Reliable evaluation of AI models is critical for scientific progress and practical application. While existing VLM benchmarks provide general insights into model capabilities, their heterogeneous designs and limited focus on a few imaging domains pose significant challenges for both cross-domain performance comparison and targeted domain-specific evaluation. To address this, we propose three key contributions: (1) a framework for the resource-efficient creation of domain-specific VLM benchmarks enabled by task augmentation for creating multiple diverse tasks from a single existing task, (2) the release of new VLM benchmarks for seven domains, created according to the same homogeneous protocol and including 162,946 thoroughly human-validated answers, and (3) an extensive benchmarking of 22 state-of-the-art VLMs on a total of 37,171 tasks, revealing performance variances across domains and tasks, thereby supporting the need for tailored VLM benchmarks. Adoption of our methodology will pave the way for the resource-efficient domain-specific selection of models and guide future research efforts toward addressing core open questions.
Confidence intervals uncovered: Are we ready for real-world medical imaging AI?
Sep 27, 2024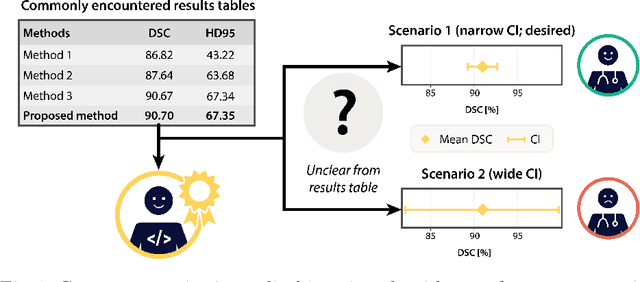
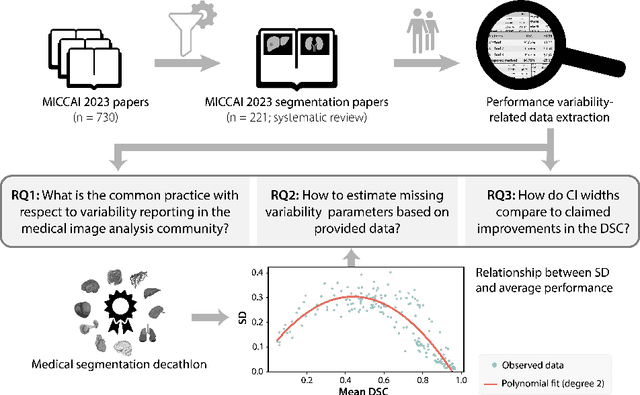
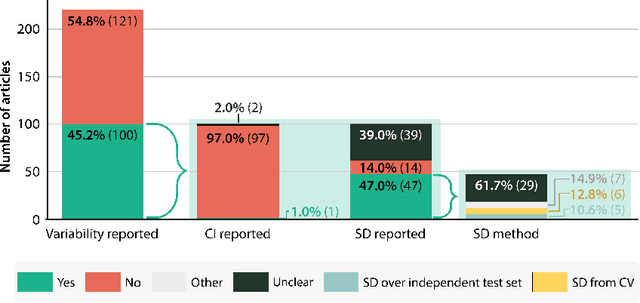

Abstract:Medical imaging is spearheading the AI transformation of healthcare. Performance reporting is key to determine which methods should be translated into clinical practice. Frequently, broad conclusions are simply derived from mean performance values. In this paper, we argue that this common practice is often a misleading simplification as it ignores performance variability. Our contribution is threefold. (1) Analyzing all MICCAI segmentation papers (n = 221) published in 2023, we first observe that more than 50% of papers do not assess performance variability at all. Moreover, only one (0.5%) paper reported confidence intervals (CIs) for model performance. (2) To address the reporting bottleneck, we show that the unreported standard deviation (SD) in segmentation papers can be approximated by a second-order polynomial function of the mean Dice similarity coefficient (DSC). Based on external validation data from 56 previous MICCAI challenges, we demonstrate that this approximation can accurately reconstruct the CI of a method using information provided in publications. (3) Finally, we reconstructed 95% CIs around the mean DSC of MICCAI 2023 segmentation papers. The median CI width was 0.03 which is three times larger than the median performance gap between the first and second ranked method. For more than 60% of papers, the mean performance of the second-ranked method was within the CI of the first-ranked method. We conclude that current publications typically do not provide sufficient evidence to support which models could potentially be translated into clinical practice.
Quality Assured: Rethinking Annotation Strategies in Imaging AI
Jul 26, 2024



Abstract:This paper does not describe a novel method. Instead, it studies an essential foundation for reliable benchmarking and ultimately real-world application of AI-based image analysis: generating high-quality reference annotations. Previous research has focused on crowdsourcing as a means of outsourcing annotations. However, little attention has so far been given to annotation companies, specifically regarding their internal quality assurance (QA) processes. Therefore, our aim is to evaluate the influence of QA employed by annotation companies on annotation quality and devise methodologies for maximizing data annotation efficacy. Based on a total of 57,648 instance segmented images obtained from a total of 924 annotators and 34 QA workers from four annotation companies and Amazon Mechanical Turk (MTurk), we derived the following insights: (1) Annotation companies perform better both in terms of quantity and quality compared to the widely used platform MTurk. (2) Annotation companies' internal QA only provides marginal improvements, if any. However, improving labeling instructions instead of investing in QA can substantially boost annotation performance. (3) The benefit of internal QA depends on specific image characteristics. Our work could enable researchers to derive substantially more value from a fixed annotation budget and change the way annotation companies conduct internal QA.
FISBe: A real-world benchmark dataset for instance segmentation of long-range thin filamentous structures
Mar 29, 2024



Abstract:Instance segmentation of neurons in volumetric light microscopy images of nervous systems enables groundbreaking research in neuroscience by facilitating joint functional and morphological analyses of neural circuits at cellular resolution. Yet said multi-neuron light microscopy data exhibits extremely challenging properties for the task of instance segmentation: Individual neurons have long-ranging, thin filamentous and widely branching morphologies, multiple neurons are tightly inter-weaved, and partial volume effects, uneven illumination and noise inherent to light microscopy severely impede local disentangling as well as long-range tracing of individual neurons. These properties reflect a current key challenge in machine learning research, namely to effectively capture long-range dependencies in the data. While respective methodological research is buzzing, to date methods are typically benchmarked on synthetic datasets. To address this gap, we release the FlyLight Instance Segmentation Benchmark (FISBe) dataset, the first publicly available multi-neuron light microscopy dataset with pixel-wise annotations. In addition, we define a set of instance segmentation metrics for benchmarking that we designed to be meaningful with regard to downstream analyses. Lastly, we provide three baselines to kick off a competition that we envision to both advance the field of machine learning regarding methodology for capturing long-range data dependencies, and facilitate scientific discovery in basic neuroscience.
Panoptica -- instance-wise evaluation of 3D semantic and instance segmentation maps
Dec 05, 2023



Abstract:This paper introduces panoptica, a versatile and performance-optimized package designed for computing instance-wise segmentation quality metrics from 2D and 3D segmentation maps. panoptica addresses the limitations of existing metrics and provides a modular framework that complements the original intersection over union-based panoptic quality with other metrics, such as the distance metric Average Symmetric Surface Distance. The package is open-source, implemented in Python, and accompanied by comprehensive documentation and tutorials. panoptica employs a three-step metrics computation process to cover diverse use cases. The efficacy of panoptica is demonstrated on various real-world biomedical datasets, where an instance-wise evaluation is instrumental for an accurate representation of the underlying clinical task. Overall, we envision panoptica as a valuable tool facilitating in-depth evaluation of segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge