Hendrik Möller
VERIDAH: Solving Enumeration Anomaly Aware Vertebra Labeling across Imaging Sequences
Jan 20, 2026Abstract:The human spine commonly consists of seven cervical, twelve thoracic, and five lumbar vertebrae. However, enumeration anomalies may result in individuals having eleven or thirteen thoracic vertebrae and four or six lumbar vertebrae. Although the identification of enumeration anomalies has potential clinical implications for chronic back pain and operation planning, the thoracolumbar junction is often poorly assessed and rarely described in clinical reports. Additionally, even though multiple deep-learning-based vertebra labeling algorithms exist, there is a lack of methods to automatically label enumeration anomalies. Our work closes that gap by introducing "Vertebra Identification with Anomaly Handling" (VERIDAH), a novel vertebra labeling algorithm based on multiple classification heads combined with a weighted vertebra sequence prediction algorithm. We show that our approach surpasses existing models on T2w TSE sagittal (98.30% vs. 94.24% of subjects with all vertebrae correctly labeled, p < 0.001) and CT imaging (99.18% vs. 77.26% of subjects with all vertebrae correctly labeled, p < 0.001) and works in arbitrary field-of-view images. VERIDAH correctly labeled the presence 2 Möller et al. of thoracic enumeration anomalies in 87.80% and 96.30% of T2w and CT images, respectively, and lumbar enumeration anomalies in 94.48% and 97.22% for T2w and CT, respectively. Our code and models are available at: https://github.com/Hendrik-code/spineps.
Rule-based Key-Point Extraction for MR-Guided Biomechanical Digital Twins of the Spine
Aug 20, 2025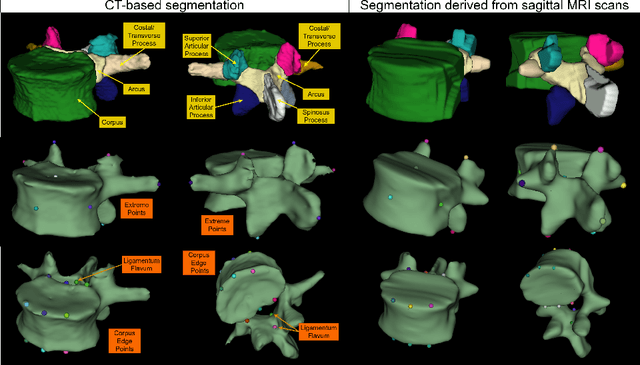

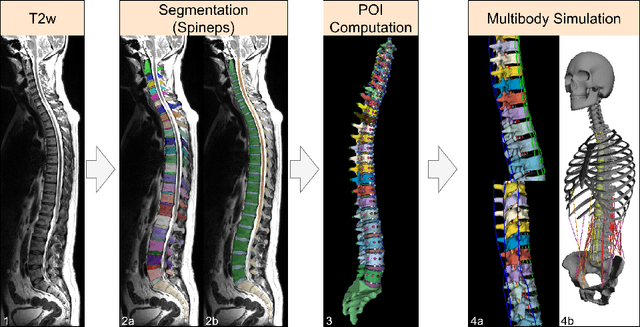

Abstract:Digital twins offer a powerful framework for subject-specific simulation and clinical decision support, yet their development often hinges on accurate, individualized anatomical modeling. In this work, we present a rule-based approach for subpixel-accurate key-point extraction from MRI, adapted from prior CT-based methods. Our approach incorporates robust image alignment and vertebra-specific orientation estimation to generate anatomically meaningful landmarks that serve as boundary conditions and force application points, like muscle and ligament insertions in biomechanical models. These models enable the simulation of spinal mechanics considering the subject's individual anatomy, and thus support the development of tailored approaches in clinical diagnostics and treatment planning. By leveraging MR imaging, our method is radiation-free and well-suited for large-scale studies and use in underrepresented populations. This work contributes to the digital twin ecosystem by bridging the gap between precise medical image analysis with biomechanical simulation, and aligns with key themes in personalized modeling for healthcare.
Automated Thoracolumbar Stump Rib Detection and Analysis in a Large CT Cohort
May 08, 2025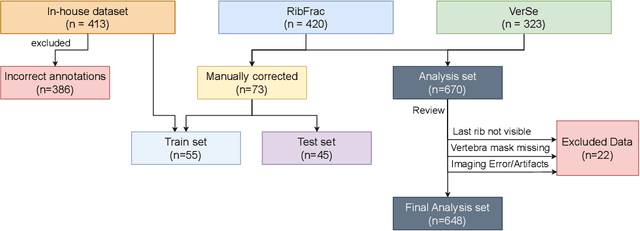
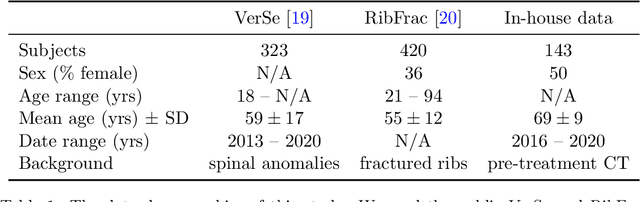
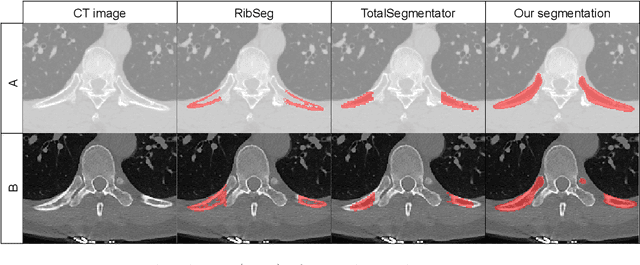
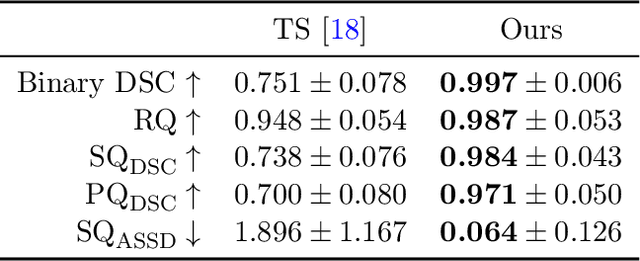
Abstract:Thoracolumbar stump ribs are one of the essential indicators of thoracolumbar transitional vertebrae or enumeration anomalies. While some studies manually assess these anomalies and describe the ribs qualitatively, this study aims to automate thoracolumbar stump rib detection and analyze their morphology quantitatively. To this end, we train a high-resolution deep-learning model for rib segmentation and show significant improvements compared to existing models (Dice score 0.997 vs. 0.779, p-value < 0.01). In addition, we use an iterative algorithm and piece-wise linear interpolation to assess the length of the ribs, showing a success rate of 98.2%. When analyzing morphological features, we show that stump ribs articulate more posteriorly at the vertebrae (-19.2 +- 3.8 vs -13.8 +- 2.5, p-value < 0.01), are thinner (260.6 +- 103.4 vs. 563.6 +- 127.1, p-value < 0.01), and are oriented more downwards and sideways within the first centimeters in contrast to full-length ribs. We show that with partially visible ribs, these features can achieve an F1-score of 0.84 in differentiating stump ribs from regular ones. We publish the model weights and masks for public use.
MAGO-SP: Detection and Correction of Water-Fat Swaps in Magnitude-Only VIBE MRI
Feb 20, 2025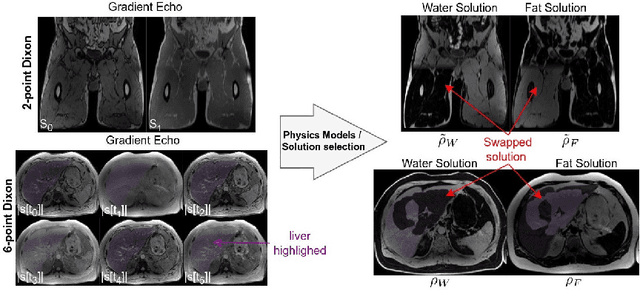
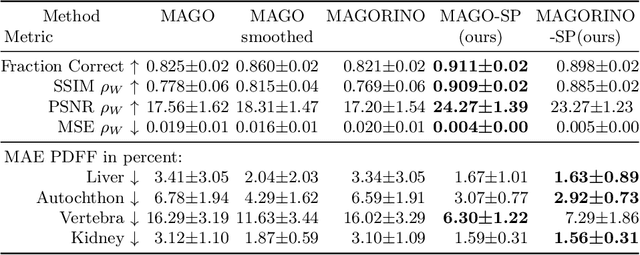
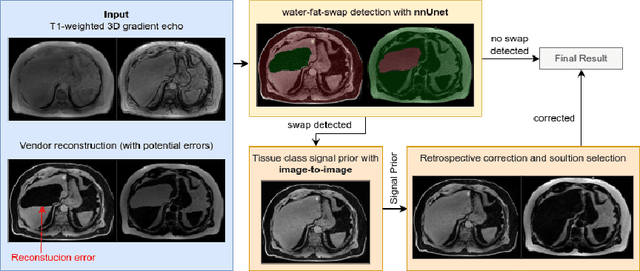
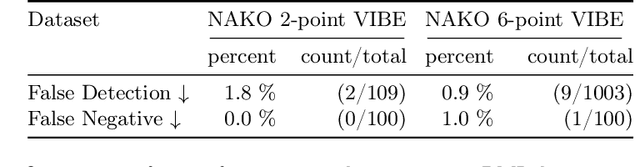
Abstract:Volume Interpolated Breath-Hold Examination (VIBE) MRI generates images suitable for water and fat signal composition estimation. While the two-point VIBE provides water-fat-separated images, the six-point VIBE allows estimation of the effective transversal relaxation rate R2* and the proton density fat fraction (PDFF), which are imaging markers for health and disease. Ambiguity during signal reconstruction can lead to water-fat swaps. This shortcoming challenges the application of VIBE-MRI for automated PDFF analyses of large-scale clinical data and of population studies. This study develops an automated pipeline to detect and correct water-fat swaps in non-contrast-enhanced VIBE images. Our three-step pipeline begins with training a segmentation network to classify volumes as "fat-like" or "water-like," using synthetic water-fat swaps generated by merging fat and water volumes with Perlin noise. Next, a denoising diffusion image-to-image network predicts water volumes as signal priors for correction. Finally, we integrate this prior into a physics-constrained model to recover accurate water and fat signals. Our approach achieves a < 1% error rate in water-fat swap detection for a 6-point VIBE. Notably, swaps disproportionately affect individuals in the Underweight and Class 3 Obesity BMI categories. Our correction algorithm ensures accurate solution selection in chemical phase MRIs, enabling reliable PDFF estimation. This forms a solid technical foundation for automated large-scale population imaging analysis.
PARASIDE: An Automatic Paranasal Sinus Segmentation and Structure Analysis Tool for MRI
Jan 24, 2025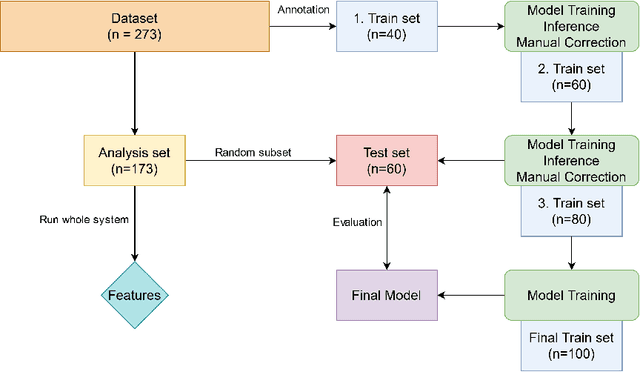
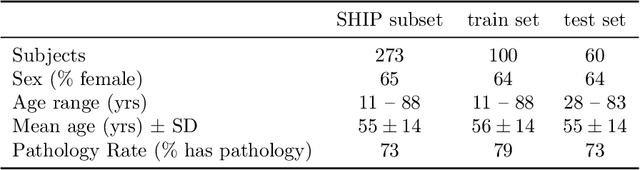
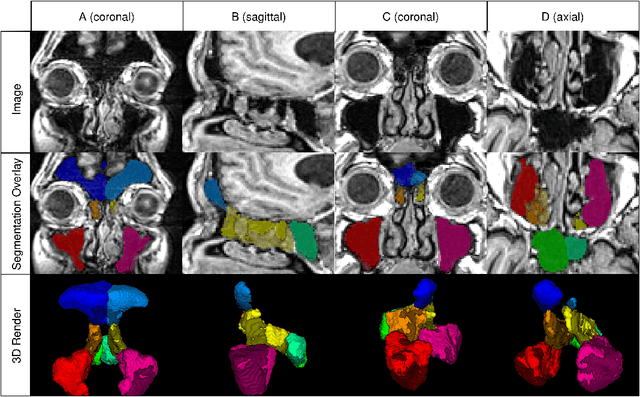
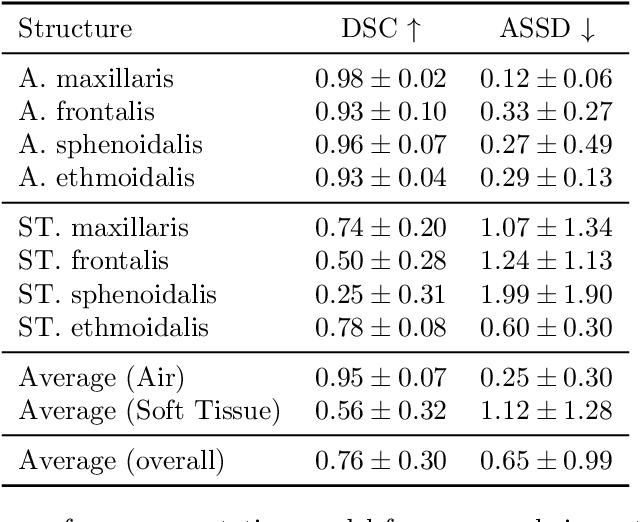
Abstract:Chronic rhinosinusitis (CRS) is a common and persistent sinus imflammation that affects 5 - 12\% of the general population. It significantly impacts quality of life and is often difficult to assess due to its subjective nature in clinical evaluation. We introduce PARASIDE, an automatic tool for segmenting air and soft tissue volumes of the structures of the sinus maxillaris, frontalis, sphenodalis and ethmoidalis in T1 MRI. By utilizing that segmentation, we can quantify feature relations that have been observed only manually and subjectively before. We performed an exemplary study and showed both volume and intensity relations between structures and radiology reports. While the soft tissue segmentation is good, the automated annotations of the air volumes are excellent. The average intensity over air structures are consistently below those of the soft tissues, close to perfect separability. Healthy subjects exhibit lower soft tissue volumes and lower intensities. Our developed system is the first automated whole nasal segmentation of 16 structures, and capable of calculating medical relevant features such as the Lund-Mackay score.
Don't You (Project Around Discs)? Neural Network Surrogate and Projected Gradient Descent for Calibrating an Intervertebral Disc Finite Element Model
Aug 12, 2024Abstract:Accurate calibration of finite element (FE) models of human intervertebral discs (IVDs) is essential for their reliability and application in diagnosing and planning treatments for spinal conditions. Traditional calibration methods are computationally intensive, requiring iterative, derivative-free optimization algorithms that often take hours or days to converge. This study addresses these challenges by introducing a novel, efficient, and effective calibration method for an L4-L5 IVD FE model using a neural network (NN) surrogate. The NN surrogate predicts simulation outcomes with high accuracy, outperforming other machine learning models, and significantly reduces the computational cost associated with traditional FE simulations. Next, a Projected Gradient Descent (PGD) approach guided by gradients of the NN surrogate is proposed to efficiently calibrate FE models. Our method explicitly enforces feasibility with a projection step, thus maintaining material bounds throughout the optimization process. The proposed method is evaluated against state-of-the-art Genetic Algorithm (GA) and inverse model baselines on synthetic and in vitro experimental datasets. Our approach demonstrates superior performance on synthetic data, achieving a Mean Absolute Error (MAE) of 0.06 compared to the baselines' MAE of 0.18 and 0.54, respectively. On experimental specimens, our method outperforms the baseline in 5 out of 6 cases. Most importantly, our approach reduces calibration time to under three seconds, compared to up to 8 days per sample required by traditional calibration. Such efficiency paves the way for applying more complex FE models, enabling accurate patient-specific simulations and advancing spinal treatment planning.
SPINEPS -- Automatic Whole Spine Segmentation of T2-weighted MR images using a Two-Phase Approach to Multi-class Semantic and Instance Segmentation
Feb 26, 2024
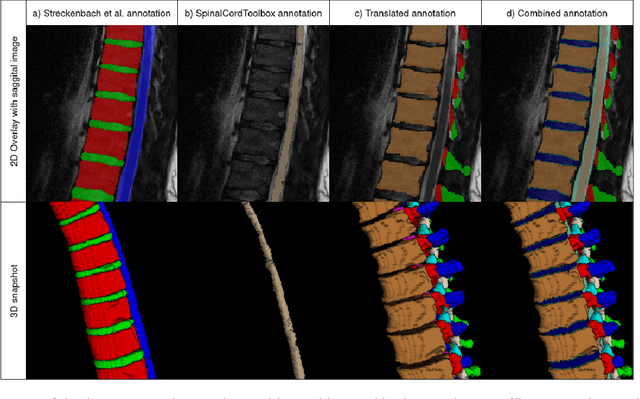
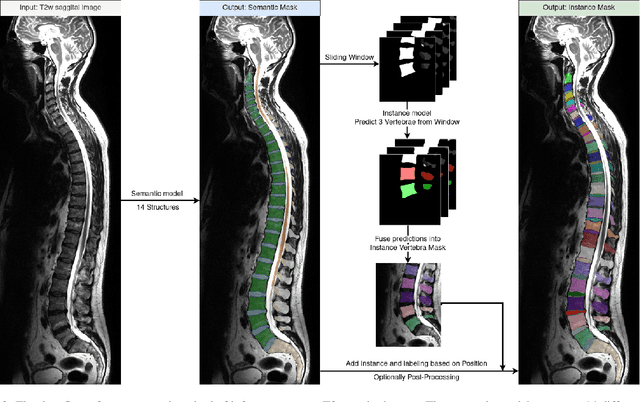
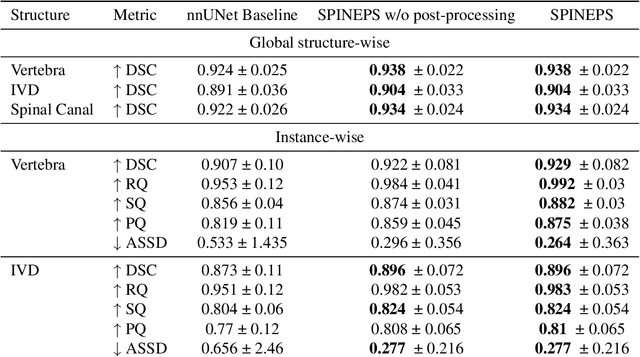
Abstract:Purpose. To present SPINEPS, an open-source deep learning approach for semantic and instance segmentation of 14 spinal structures (ten vertebra substructures, intervertebral discs, spinal cord, spinal canal, and sacrum) in whole body T2w MRI. Methods. During this HIPPA-compliant, retrospective study, we utilized the public SPIDER dataset (218 subjects, 63% female) and a subset of the German National Cohort (1423 subjects, mean age 53, 49% female) for training and evaluation. We combined CT and T2w segmentations to train models that segment 14 spinal structures in T2w sagittal scans both semantically and instance-wise. Performance evaluation metrics included Dice similarity coefficient, average symmetrical surface distance, panoptic quality, segmentation quality, and recognition quality. Statistical significance was assessed using the Wilcoxon signed-rank test. An in-house dataset was used to qualitatively evaluate out-of-distribution samples. Results. On the public dataset, our approach outperformed the baseline (instance-wise vertebra dice score 0.929 vs. 0.907, p-value<0.001). Training on auto-generated annotations and evaluating on manually corrected test data from the GNC yielded global dice scores of 0.900 for vertebrae, 0.960 for intervertebral discs, and 0.947 for the spinal canal. Incorporating the SPIDER dataset during training increased these scores to 0.920, 0.967, 0.958, respectively. Conclusions. The proposed segmentation approach offers robust segmentation of 14 spinal structures in T2w sagittal images, including the spinal cord, spinal canal, intervertebral discs, endplate, sacrum, and vertebrae. The approach yields both a semantic and instance mask as output, thus being easy to utilize. This marks the first publicly available algorithm for whole spine segmentation in sagittal T2w MR imaging.
Panoptica -- instance-wise evaluation of 3D semantic and instance segmentation maps
Dec 05, 2023



Abstract:This paper introduces panoptica, a versatile and performance-optimized package designed for computing instance-wise segmentation quality metrics from 2D and 3D segmentation maps. panoptica addresses the limitations of existing metrics and provides a modular framework that complements the original intersection over union-based panoptic quality with other metrics, such as the distance metric Average Symmetric Surface Distance. The package is open-source, implemented in Python, and accompanied by comprehensive documentation and tutorials. panoptica employs a three-step metrics computation process to cover diverse use cases. The efficacy of panoptica is demonstrated on various real-world biomedical datasets, where an instance-wise evaluation is instrumental for an accurate representation of the underlying clinical task. Overall, we envision panoptica as a valuable tool facilitating in-depth evaluation of segmentation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge