Annette Peters
MAGO-SP: Detection and Correction of Water-Fat Swaps in Magnitude-Only VIBE MRI
Feb 20, 2025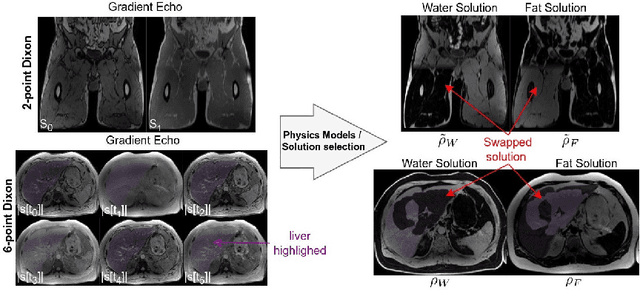
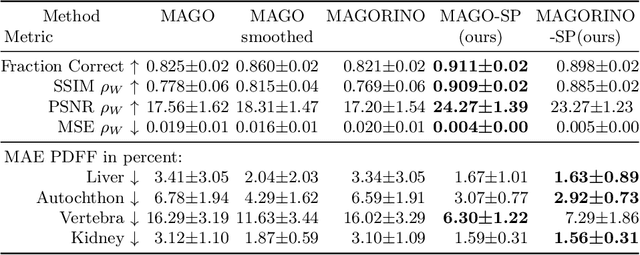
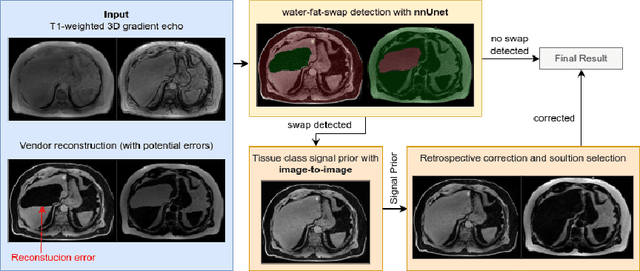
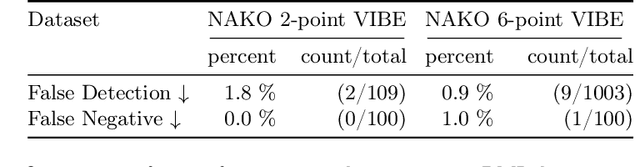
Abstract:Volume Interpolated Breath-Hold Examination (VIBE) MRI generates images suitable for water and fat signal composition estimation. While the two-point VIBE provides water-fat-separated images, the six-point VIBE allows estimation of the effective transversal relaxation rate R2* and the proton density fat fraction (PDFF), which are imaging markers for health and disease. Ambiguity during signal reconstruction can lead to water-fat swaps. This shortcoming challenges the application of VIBE-MRI for automated PDFF analyses of large-scale clinical data and of population studies. This study develops an automated pipeline to detect and correct water-fat swaps in non-contrast-enhanced VIBE images. Our three-step pipeline begins with training a segmentation network to classify volumes as "fat-like" or "water-like," using synthetic water-fat swaps generated by merging fat and water volumes with Perlin noise. Next, a denoising diffusion image-to-image network predicts water volumes as signal priors for correction. Finally, we integrate this prior into a physics-constrained model to recover accurate water and fat signals. Our approach achieves a < 1% error rate in water-fat swap detection for a 6-point VIBE. Notably, swaps disproportionately affect individuals in the Underweight and Class 3 Obesity BMI categories. Our correction algorithm ensures accurate solution selection in chemical phase MRIs, enabling reliable PDFF estimation. This forms a solid technical foundation for automated large-scale population imaging analysis.
FedNorm: Modality-Based Normalization in Federated Learning for Multi-Modal Liver Segmentation
May 23, 2022



Abstract:Given the high incidence and effective treatment options for liver diseases, they are of great socioeconomic importance. One of the most common methods for analyzing CT and MRI images for diagnosis and follow-up treatment is liver segmentation. Recent advances in deep learning have demonstrated encouraging results for automatic liver segmentation. Despite this, their success depends primarily on the availability of an annotated database, which is often not available because of privacy concerns. Federated Learning has been recently proposed as a solution to alleviate these challenges by training a shared global model on distributed clients without access to their local databases. Nevertheless, Federated Learning does not perform well when it is trained on a high degree of heterogeneity of image data due to multi-modal imaging, such as CT and MRI, and multiple scanner types. To this end, we propose Fednorm and its extension \fednormp, two Federated Learning algorithms that use a modality-based normalization technique. Specifically, Fednorm normalizes the features on a client-level, while Fednorm+ employs the modality information of single slices in the feature normalization. Our methods were validated using 428 patients from six publicly available databases and compared to state-of-the-art Federated Learning algorithms and baseline models in heterogeneous settings (multi-institutional, multi-modal data). The experimental results demonstrate that our methods show an overall acceptable performance, achieve Dice per patient scores up to 0.961, consistently outperform locally trained models, and are on par or slightly better than centralized models.
Predicting brain-age from raw T 1 -weighted Magnetic Resonance Imaging data using 3D Convolutional Neural Networks
Mar 22, 2021



Abstract:Age prediction based on Magnetic Resonance Imaging (MRI) data of the brain is a biomarker to quantify the progress of brain diseases and aging. Current approaches rely on preparing the data with multiple preprocessing steps, such as registering voxels to a standardized brain atlas, which yields a significant computational overhead, hampers widespread usage and results in the predicted brain-age to be sensitive to preprocessing parameters. Here we describe a 3D Convolutional Neural Network (CNN) based on the ResNet architecture being trained on raw, non-registered T$_ 1$-weighted MRI data of N=10,691 samples from the German National Cohort and additionally applied and validated in N=2,173 samples from three independent studies using transfer learning. For comparison, state-of-the-art models using preprocessed neuroimaging data are trained and validated on the same samples. The 3D CNN using raw neuroimaging data predicts age with a mean average deviation of 2.84 years, outperforming the state-of-the-art brain-age models using preprocessed data. Since our approach is invariant to preprocessing software and parameter choices, it enables faster, more robust and more accurate brain-age modeling.
Deep Shape Analysis on Abdominal Organs for Diabetes Prediction
Aug 06, 2018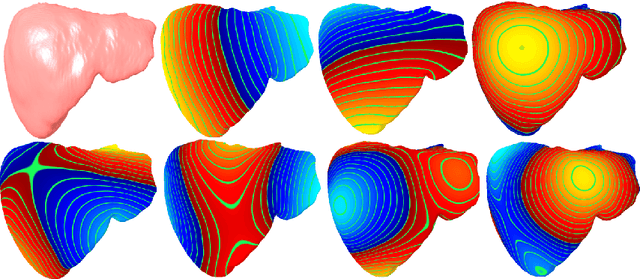
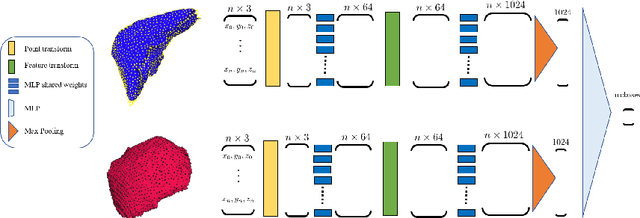
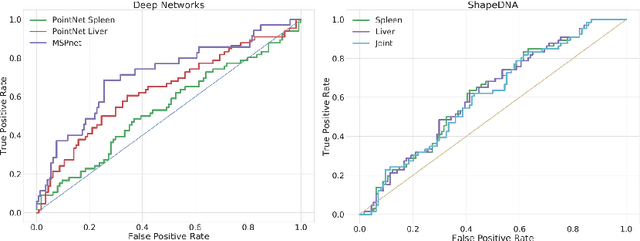
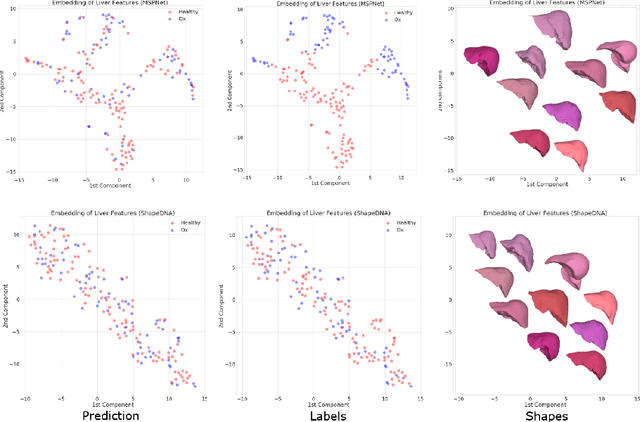
Abstract:Morphological analysis of organs based on images is a key task in medical imaging computing. Several approaches have been proposed for the quantitative assessment of morphological changes, and they have been widely used for the analysis of the effects of aging, disease and other factors in organ morphology. In this work, we propose a deep neural network for predicting diabetes on abdominal shapes. The network directly operates on raw point clouds without requiring mesh processing or shape alignment. Instead of relying on hand-crafted shape descriptors, an optimal representation is learned in the end-to-end training stage of the network. For comparison, we extend the state-of-the-art shape descriptor BrainPrint to the AbdomenPrint. Our results demonstrate that the network learns shape representations that better separates healthy and diabetic individuals than traditional representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge