Shadi Albarqouni
University Hospital Bonn, Venusberg-Campus 1, D-53127, Bonn, Germany, Helmholtz Munich, Ingolstädter Landstraße 1, D-85764, Neuherberg, Germany, Technical University of Munich, Boltzmannstr. 3, D-85748 Garching, Germany
INTERACT-CMIL: Multi-Task Shared Learning and Inter-Task Consistency for Conjunctival Melanocytic Intraepithelial Lesion Grading
Dec 27, 2025Abstract:Accurate grading of Conjunctival Melanocytic Intraepithelial Lesions (CMIL) is essential for treatment and melanoma prediction but remains difficult due to subtle morphological cues and interrelated diagnostic criteria. We introduce INTERACT-CMIL, a multi-head deep learning framework that jointly predicts five histopathological axes; WHO4, WHO5, horizontal spread, vertical spread, and cytologic atypia, through Shared Feature Learning with Combinatorial Partial Supervision and an Inter-Dependence Loss enforcing cross-task consistency. Trained and evaluated on a newly curated, multi-center dataset of 486 expert-annotated conjunctival biopsy patches from three university hospitals, INTERACT-CMIL achieves consistent improvements over CNN and foundation-model (FM) baselines, with relative macro F1 gains up to 55.1% (WHO4) and 25.0% (vertical spread). The framework provides coherent, interpretable multi-criteria predictions aligned with expert grading, offering a reproducible computational benchmark for CMIL diagnosis and a step toward standardized digital ocular pathology.
MRIQT: Physics-Aware Diffusion Model for Image Quality Transfer in Neonatal Ultra-Low-Field MRI
Nov 17, 2025Abstract:Portable ultra-low-field MRI (uLF-MRI, 0.064 T) offers accessible neuroimaging for neonatal care but suffers from low signal-to-noise ratio and poor diagnostic quality compared to high-field (HF) MRI. We propose MRIQT, a 3D conditional diffusion framework for image quality transfer (IQT) from uLF to HF MRI. MRIQT combines realistic K-space degradation for physics-consistent uLF simulation, v-prediction with classifier-free guidance for stable image-to-image generation, and an SNR-weighted 3D perceptual loss for anatomical fidelity. The model denoises from a noised uLF input conditioned on the same scan, leveraging volumetric attention-UNet architecture for structure-preserving translation. Trained on a neonatal cohort with diverse pathologies, MRIQT surpasses recent GAN and CNN baselines in PSNR 15.3% with 1.78% over the state of the art, while physicians rated 85% of its outputs as good quality with clear pathology present. MRIQT enables high-fidelity, diffusion-based enhancement of portable ultra-low-field (uLF) MRI for deliable neonatal brain assessment.
ProSona: Prompt-Guided Personalization for Multi-Expert Medical Image Segmentation
Nov 11, 2025Abstract:Automated medical image segmentation suffers from high inter-observer variability, particularly in tasks such as lung nodule delineation, where experts often disagree. Existing approaches either collapse this variability into a consensus mask or rely on separate model branches for each annotator. We introduce ProSona, a two-stage framework that learns a continuous latent space of annotation styles, enabling controllable personalization via natural language prompts. A probabilistic U-Net backbone captures diverse expert hypotheses, while a prompt-guided projection mechanism navigates this latent space to generate personalized segmentations. A multi-level contrastive objective aligns textual and visual representations, promoting disentangled and interpretable expert styles. Across the LIDC-IDRI lung nodule and multi-institutional prostate MRI datasets, ProSona reduces the Generalized Energy Distance by 17% and improves mean Dice by more than one point compared with DPersona. These results demonstrate that natural-language prompts can provide flexible, accurate, and interpretable control over personalized medical image segmentation. Our implementation is available online 1 .
Bias and Generalizability of Foundation Models across Datasets in Breast Mammography
May 19, 2025Abstract:Over the past decades, computer-aided diagnosis tools for breast cancer have been developed to enhance screening procedures, yet their clinical adoption remains challenged by data variability and inherent biases. Although foundation models (FMs) have recently demonstrated impressive generalizability and transfer learning capabilities by leveraging vast and diverse datasets, their performance can be undermined by spurious correlations that arise from variations in image quality, labeling uncertainty, and sensitive patient attributes. In this work, we explore the fairness and bias of FMs for breast mammography classification by leveraging a large pool of datasets from diverse sources-including data from underrepresented regions and an in-house dataset. Our extensive experiments show that while modality-specific pre-training of FMs enhances performance, classifiers trained on features from individual datasets fail to generalize across domains. Aggregating datasets improves overall performance, yet does not fully mitigate biases, leading to significant disparities across under-represented subgroups such as extreme breast densities and age groups. Furthermore, while domain-adaptation strategies can reduce these disparities, they often incur a performance trade-off. In contrast, fairness-aware techniques yield more stable and equitable performance across subgroups. These findings underscore the necessity of incorporating rigorous fairness evaluations and mitigation strategies into FM-based models to foster inclusive and generalizable AI.
LVM-Med: Learning Large-Scale Self-Supervised Vision Models for Medical Imaging via Second-order Graph Matching
Jul 09, 2023



Abstract:Obtaining large pre-trained models that can be fine-tuned to new tasks with limited annotated samples has remained an open challenge for medical imaging data. While pre-trained deep networks on ImageNet and vision-language foundation models trained on web-scale data are prevailing approaches, their effectiveness on medical tasks is limited due to the significant domain shift between natural and medical images. To bridge this gap, we introduce LVM-Med, the first family of deep networks trained on large-scale medical datasets. We have collected approximately 1.3 million medical images from 55 publicly available datasets, covering a large number of organs and modalities such as CT, MRI, X-ray, and Ultrasound. We benchmark several state-of-the-art self-supervised algorithms on this dataset and propose a novel self-supervised contrastive learning algorithm using a graph-matching formulation. The proposed approach makes three contributions: (i) it integrates prior pair-wise image similarity metrics based on local and global information; (ii) it captures the structural constraints of feature embeddings through a loss function constructed via a combinatorial graph-matching objective; and (iii) it can be trained efficiently end-to-end using modern gradient-estimation techniques for black-box solvers. We thoroughly evaluate the proposed LVM-Med on 15 downstream medical tasks ranging from segmentation and classification to object detection, and both for the in and out-of-distribution settings. LVM-Med empirically outperforms a number of state-of-the-art supervised, self-supervised, and foundation models. For challenging tasks such as Brain Tumor Classification or Diabetic Retinopathy Grading, LVM-Med improves previous vision-language models trained on 1 billion masks by 6-7% while using only a ResNet-50.
LYSTO: The Lymphocyte Assessment Hackathon and Benchmark Dataset
Jan 16, 2023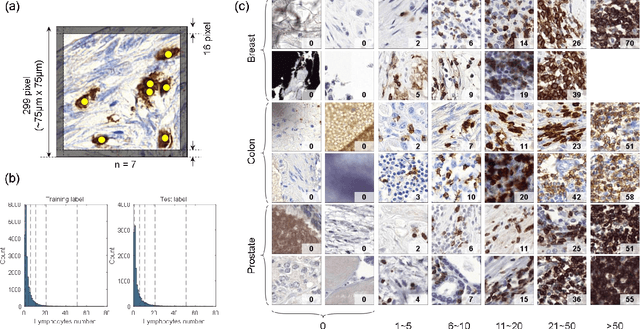
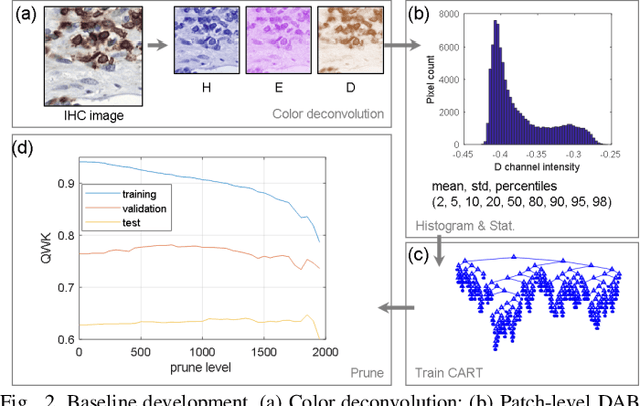
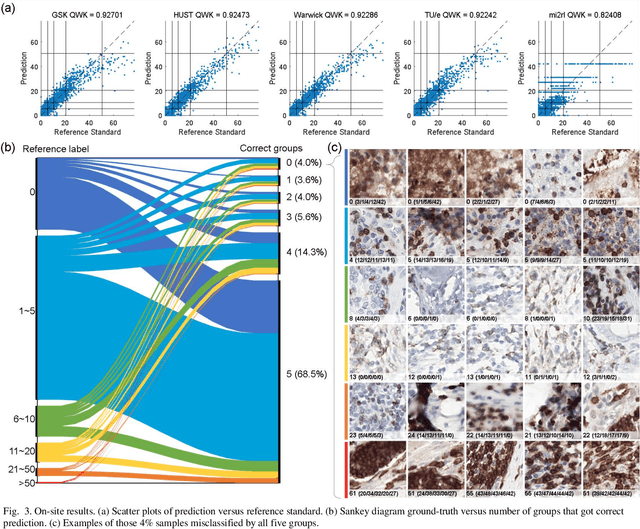
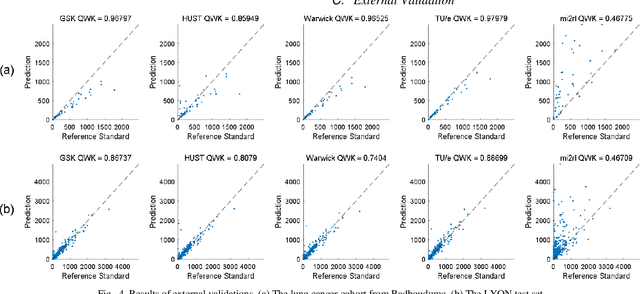
Abstract:We introduce LYSTO, the Lymphocyte Assessment Hackathon, which was held in conjunction with the MICCAI 2019 Conference in Shenzen (China). The competition required participants to automatically assess the number of lymphocytes, in particular T-cells, in histopathological images of colon, breast, and prostate cancer stained with CD3 and CD8 immunohistochemistry. Differently from other challenges setup in medical image analysis, LYSTO participants were solely given a few hours to address this problem. In this paper, we describe the goal and the multi-phase organization of the hackathon; we describe the proposed methods and the on-site results. Additionally, we present post-competition results where we show how the presented methods perform on an independent set of lung cancer slides, which was not part of the initial competition, as well as a comparison on lymphocyte assessment between presented methods and a panel of pathologists. We show that some of the participants were capable to achieve pathologist-level performance at lymphocyte assessment. After the hackathon, LYSTO was left as a lightweight plug-and-play benchmark dataset on grand-challenge website, together with an automatic evaluation platform. LYSTO has supported a number of research in lymphocyte assessment in oncology. LYSTO will be a long-lasting educational challenge for deep learning and digital pathology, it is available at https://lysto.grand-challenge.org/.
Joint Self-Supervised Image-Volume Representation Learning with Intra-Inter Contrastive Clustering
Dec 04, 2022



Abstract:Collecting large-scale medical datasets with fully annotated samples for training of deep networks is prohibitively expensive, especially for 3D volume data. Recent breakthroughs in self-supervised learning (SSL) offer the ability to overcome the lack of labeled training samples by learning feature representations from unlabeled data. However, most current SSL techniques in the medical field have been designed for either 2D images or 3D volumes. In practice, this restricts the capability to fully leverage unlabeled data from numerous sources, which may include both 2D and 3D data. Additionally, the use of these pre-trained networks is constrained to downstream tasks with compatible data dimensions. In this paper, we propose a novel framework for unsupervised joint learning on 2D and 3D data modalities. Given a set of 2D images or 2D slices extracted from 3D volumes, we construct an SSL task based on a 2D contrastive clustering problem for distinct classes. The 3D volumes are exploited by computing vectored embedding at each slice and then assembling a holistic feature through deformable self-attention mechanisms in Transformer, allowing incorporating long-range dependencies between slices inside 3D volumes. These holistic features are further utilized to define a novel 3D clustering agreement-based SSL task and masking embedding prediction inspired by pre-trained language models. Experiments on downstream tasks, such as 3D brain segmentation, lung nodule detection, 3D heart structures segmentation, and abnormal chest X-ray detection, demonstrate the effectiveness of our joint 2D and 3D SSL approach. We improve plain 2D Deep-ClusterV2 and SwAV by a significant margin and also surpass various modern 2D and 3D SSL approaches.
What can we learn about a generated image corrupting its latent representation?
Oct 12, 2022Abstract:Generative adversarial networks (GANs) offer an effective solution to the image-to-image translation problem, thereby allowing for new possibilities in medical imaging. They can translate images from one imaging modality to another at a low cost. For unpaired datasets, they rely mostly on cycle loss. Despite its effectiveness in learning the underlying data distribution, it can lead to a discrepancy between input and output data. The purpose of this work is to investigate the hypothesis that we can predict image quality based on its latent representation in the GANs bottleneck. We achieve this by corrupting the latent representation with noise and generating multiple outputs. The degree of differences between them is interpreted as the strength of the representation: the more robust the latent representation, the fewer changes in the output image the corruption causes. Our results demonstrate that our proposed method has the ability to i) predict uncertain parts of synthesized images, and ii) identify samples that may not be reliable for downstream tasks, e.g., liver segmentation task.
FLamby: Datasets and Benchmarks for Cross-Silo Federated Learning in Realistic Healthcare Settings
Oct 10, 2022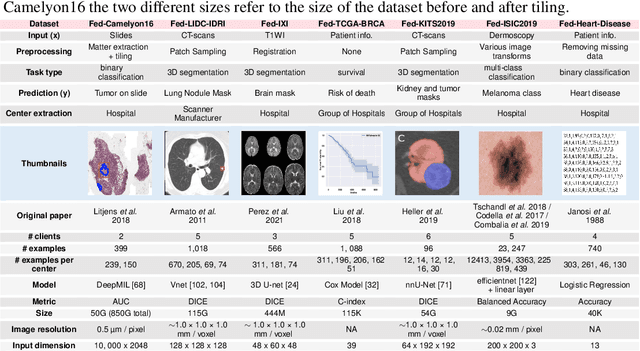
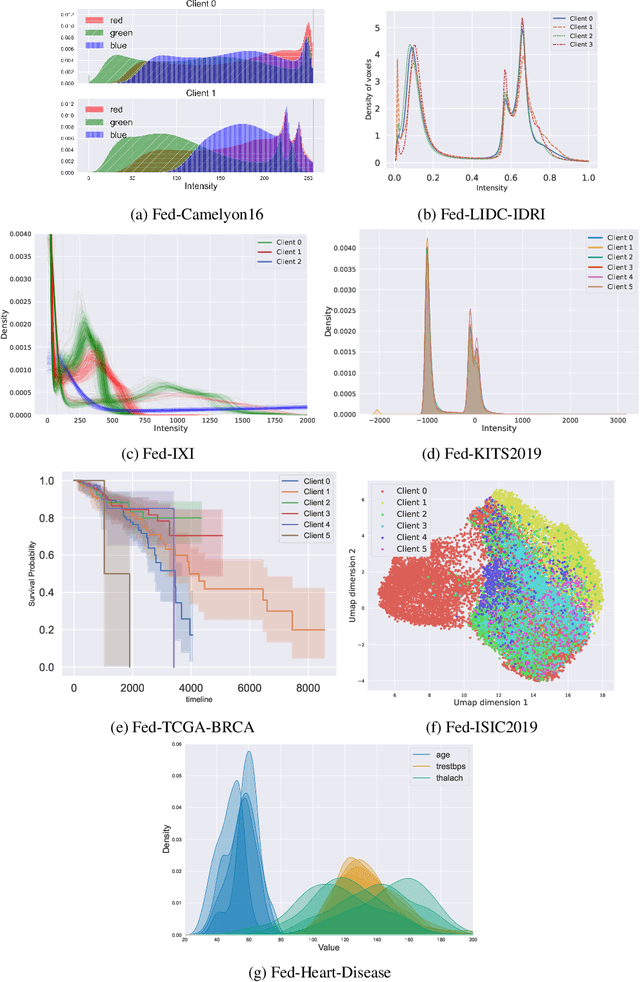
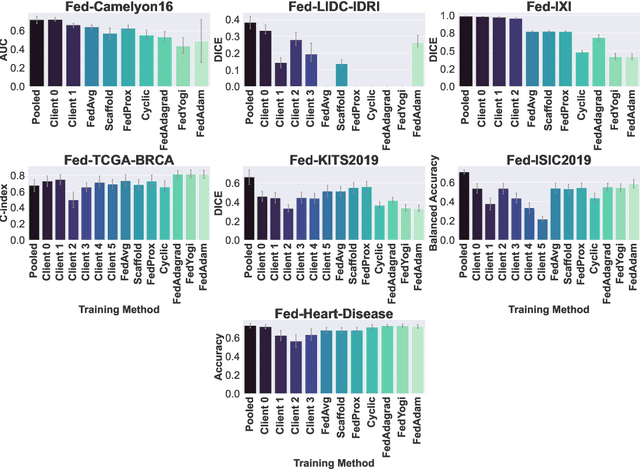
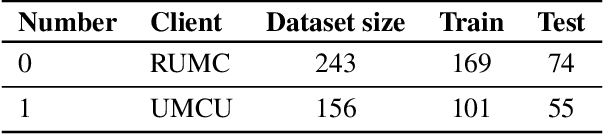
Abstract:Federated Learning (FL) is a novel approach enabling several clients holding sensitive data to collaboratively train machine learning models, without centralizing data. The cross-silo FL setting corresponds to the case of few ($2$--$50$) reliable clients, each holding medium to large datasets, and is typically found in applications such as healthcare, finance, or industry. While previous works have proposed representative datasets for cross-device FL, few realistic healthcare cross-silo FL datasets exist, thereby slowing algorithmic research in this critical application. In this work, we propose a novel cross-silo dataset suite focused on healthcare, FLamby (Federated Learning AMple Benchmark of Your cross-silo strategies), to bridge the gap between theory and practice of cross-silo FL. FLamby encompasses 7 healthcare datasets with natural splits, covering multiple tasks, modalities, and data volumes, each accompanied with baseline training code. As an illustration, we additionally benchmark standard FL algorithms on all datasets. Our flexible and modular suite allows researchers to easily download datasets, reproduce results and re-use the different components for their research. FLamby is available at~\url{www.github.com/owkin/flamby}.
Transformer based Models for Unsupervised Anomaly Segmentation in Brain MR Images
Jul 05, 2022
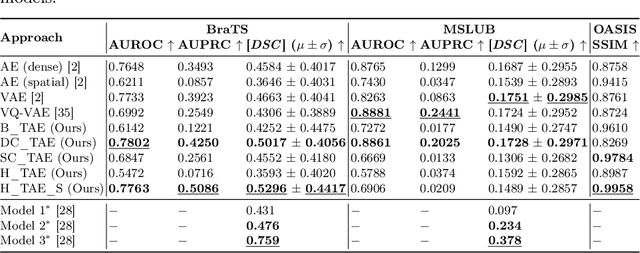


Abstract:The quality of patient care associated with diagnostic radiology is proportionate to a physician workload. Segmentation is a fundamental limiting precursor to diagnostic and therapeutic procedures. Advances in Machine Learning (ML) aim to increase diagnostic efficiency to replace single application with generalized algorithms. In Unsupervised Anomaly Detection (UAD), Convolutional Neural Network (CNN) based Autoencoders (AEs) and Variational Autoencoders (VAEs) are considered as a de facto approach for reconstruction based anomaly segmentation. Looking for anomalous regions in medical images is one of the main applications that use anomaly segmentation. The restricted receptive field in CNNs limit the CNN to model the global context and hence if the anomalous regions cover parts of the image, the CNN-based AEs are not capable to bring semantic understanding of the image. On the other hand, Vision Transformers (ViTs) have emerged as a competitive alternative to CNNs. It relies on the self-attention mechanism that is capable to relate image patches to each other. To reconstruct a coherent and more realistic image, in this work, we investigate Transformer capabilities in building AEs for reconstruction based UAD task. We focus on anomaly segmentation for Brain Magnetic Resonance Imaging (MRI) and present five Transformer-based models while enabling segmentation performance comparable or superior to State-of-The-Art (SOTA) models. The source code is available on Github https://github.com/ahmedgh970/Transformers_Unsupervised_Anomaly_Segmentation.git
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge