Mai T. N. Truong
DRG-Net: Interactive Joint Learning of Multi-lesion Segmentation and Classification for Diabetic Retinopathy Grading
Dec 30, 2022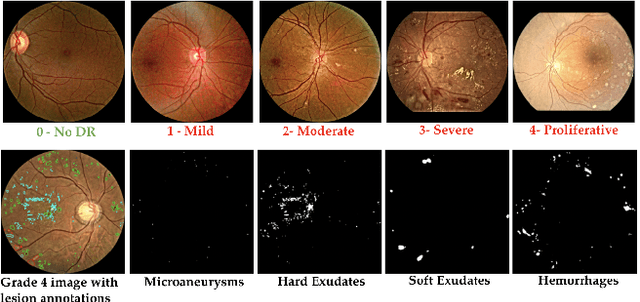
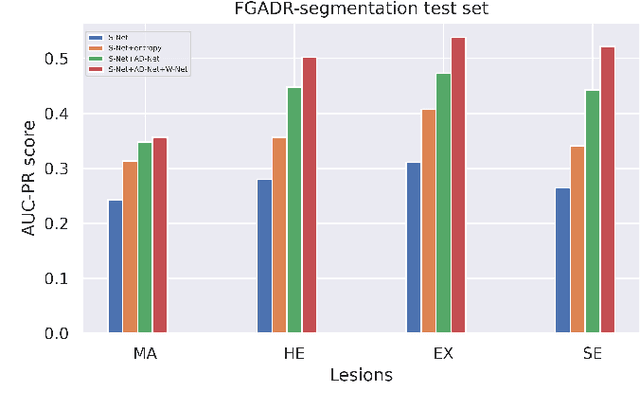
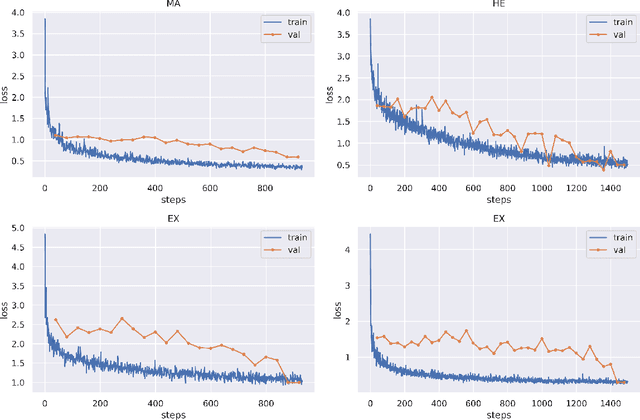
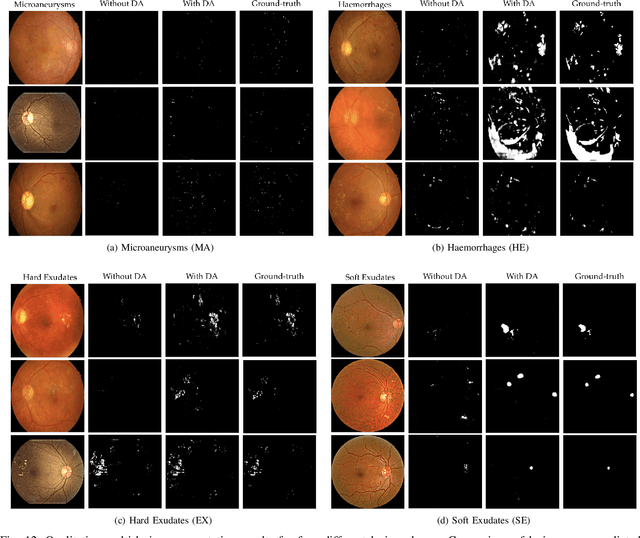
Abstract:Diabetic Retinopathy (DR) is a leading cause of vision loss in the world, and early DR detection is necessary to prevent vision loss and support an appropriate treatment. In this work, we leverage interactive machine learning and introduce a joint learning framework, termed DRG-Net, to effectively learn both disease grading and multi-lesion segmentation. Our DRG-Net consists of two modules: (i) DRG-AI-System to classify DR Grading, localize lesion areas, and provide visual explanations; (ii) DRG-Expert-Interaction to receive feedback from user-expert and improve the DRG-AI-System. To deal with sparse data, we utilize transfer learning mechanisms to extract invariant feature representations by using Wasserstein distance and adversarial learning-based entropy minimization. Besides, we propose a novel attention strategy at both low- and high-level features to automatically select the most significant lesion information and provide explainable properties. In terms of human interaction, we further develop DRG-Net as a tool that enables expert users to correct the system's predictions, which may then be used to update the system as a whole. Moreover, thanks to the attention mechanism and loss functions constraint between lesion features and classification features, our approach can be robust given a certain level of noise in the feedback of users. We have benchmarked DRG-Net on the two largest DR datasets, i.e., IDRID and FGADR, and compared it to various state-of-the-art deep learning networks. In addition to outperforming other SOTA approaches, DRG-Net is effectively updated using user feedback, even in a weakly-supervised manner.
Joint Self-Supervised Image-Volume Representation Learning with Intra-Inter Contrastive Clustering
Dec 04, 2022



Abstract:Collecting large-scale medical datasets with fully annotated samples for training of deep networks is prohibitively expensive, especially for 3D volume data. Recent breakthroughs in self-supervised learning (SSL) offer the ability to overcome the lack of labeled training samples by learning feature representations from unlabeled data. However, most current SSL techniques in the medical field have been designed for either 2D images or 3D volumes. In practice, this restricts the capability to fully leverage unlabeled data from numerous sources, which may include both 2D and 3D data. Additionally, the use of these pre-trained networks is constrained to downstream tasks with compatible data dimensions. In this paper, we propose a novel framework for unsupervised joint learning on 2D and 3D data modalities. Given a set of 2D images or 2D slices extracted from 3D volumes, we construct an SSL task based on a 2D contrastive clustering problem for distinct classes. The 3D volumes are exploited by computing vectored embedding at each slice and then assembling a holistic feature through deformable self-attention mechanisms in Transformer, allowing incorporating long-range dependencies between slices inside 3D volumes. These holistic features are further utilized to define a novel 3D clustering agreement-based SSL task and masking embedding prediction inspired by pre-trained language models. Experiments on downstream tasks, such as 3D brain segmentation, lung nodule detection, 3D heart structures segmentation, and abnormal chest X-ray detection, demonstrate the effectiveness of our joint 2D and 3D SSL approach. We improve plain 2D Deep-ClusterV2 and SwAV by a significant margin and also surpass various modern 2D and 3D SSL approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge