Tri Cao
Just-In-Time Reinforcement Learning: Continual Learning in LLM Agents Without Gradient Updates
Jan 26, 2026Abstract:While Large Language Model (LLM) agents excel at general tasks, they inherently struggle with continual adaptation due to the frozen weights after deployment. Conventional reinforcement learning (RL) offers a solution but incurs prohibitive computational costs and the risk of catastrophic forgetting. We introduce Just-In-Time Reinforcement Learning (JitRL), a training-free framework that enables test-time policy optimization without any gradient updates. JitRL maintains a dynamic, non-parametric memory of experiences and retrieves relevant trajectories to estimate action advantages on-the-fly. These estimates are then used to directly modulate the LLM's output logits. We theoretically prove that this additive update rule is the exact closed-form solution to the KL-constrained policy optimization objective. Extensive experiments on WebArena and Jericho demonstrate that JitRL establishes a new state-of-the-art among training-free methods. Crucially, JitRL outperforms the performance of computationally expensive fine-tuning methods (e.g., WebRL) while reducing monetary costs by over 30 times, offering a scalable path for continual learning agents. The code is available at https://github.com/liushiliushi/JitRL.
Automating Steering for Safe Multimodal Large Language Models
Jul 17, 2025Abstract:Recent progress in Multimodal Large Language Models (MLLMs) has unlocked powerful cross-modal reasoning abilities, but also raised new safety concerns, particularly when faced with adversarial multimodal inputs. To improve the safety of MLLMs during inference, we introduce a modular and adaptive inference-time intervention technology, AutoSteer, without requiring any fine-tuning of the underlying model. AutoSteer incorporates three core components: (1) a novel Safety Awareness Score (SAS) that automatically identifies the most safety-relevant distinctions among the model's internal layers; (2) an adaptive safety prober trained to estimate the likelihood of toxic outputs from intermediate representations; and (3) a lightweight Refusal Head that selectively intervenes to modulate generation when safety risks are detected. Experiments on LLaVA-OV and Chameleon across diverse safety-critical benchmarks demonstrate that AutoSteer significantly reduces the Attack Success Rate (ASR) for textual, visual, and cross-modal threats, while maintaining general abilities. These findings position AutoSteer as a practical, interpretable, and effective framework for safer deployment of multimodal AI systems.
GuardReasoner-VL: Safeguarding VLMs via Reinforced Reasoning
May 16, 2025Abstract:To enhance the safety of VLMs, this paper introduces a novel reasoning-based VLM guard model dubbed GuardReasoner-VL. The core idea is to incentivize the guard model to deliberatively reason before making moderation decisions via online RL. First, we construct GuardReasoner-VLTrain, a reasoning corpus with 123K samples and 631K reasoning steps, spanning text, image, and text-image inputs. Then, based on it, we cold-start our model's reasoning ability via SFT. In addition, we further enhance reasoning regarding moderation through online RL. Concretely, to enhance diversity and difficulty of samples, we conduct rejection sampling followed by data augmentation via the proposed safety-aware data concatenation. Besides, we use a dynamic clipping parameter to encourage exploration in early stages and exploitation in later stages. To balance performance and token efficiency, we design a length-aware safety reward that integrates accuracy, format, and token cost. Extensive experiments demonstrate the superiority of our model. Remarkably, it surpasses the runner-up by 19.27% F1 score on average. We release data, code, and models (3B/7B) of GuardReasoner-VL at https://github.com/yueliu1999/GuardReasoner-VL/
Words or Vision: Do Vision-Language Models Have Blind Faith in Text?
Mar 04, 2025Abstract:Vision-Language Models (VLMs) excel in integrating visual and textual information for vision-centric tasks, but their handling of inconsistencies between modalities is underexplored. We investigate VLMs' modality preferences when faced with visual data and varied textual inputs in vision-centered settings. By introducing textual variations to four vision-centric tasks and evaluating ten Vision-Language Models (VLMs), we discover a \emph{``blind faith in text''} phenomenon: VLMs disproportionately trust textual data over visual data when inconsistencies arise, leading to significant performance drops under corrupted text and raising safety concerns. We analyze factors influencing this text bias, including instruction prompts, language model size, text relevance, token order, and the interplay between visual and textual certainty. While certain factors, such as scaling up the language model size, slightly mitigate text bias, others like token order can exacerbate it due to positional biases inherited from language models. To address this issue, we explore supervised fine-tuning with text augmentation and demonstrate its effectiveness in reducing text bias. Additionally, we provide a theoretical analysis suggesting that the blind faith in text phenomenon may stem from an imbalance of pure text and multi-modal data during training. Our findings highlight the need for balanced training and careful consideration of modality interactions in VLMs to enhance their robustness and reliability in handling multi-modal data inconsistencies.
Meta-Reasoner: Dynamic Guidance for Optimized Inference-time Reasoning in Large Language Models
Feb 27, 2025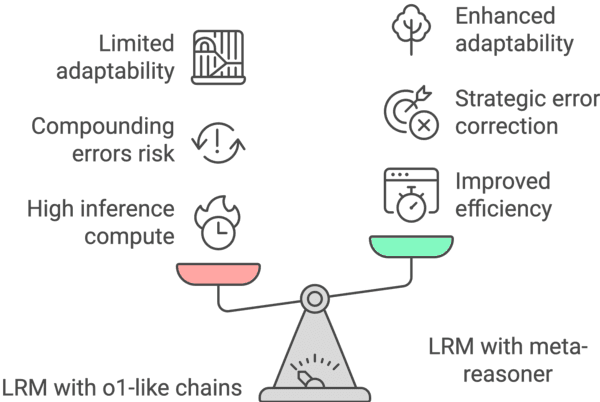
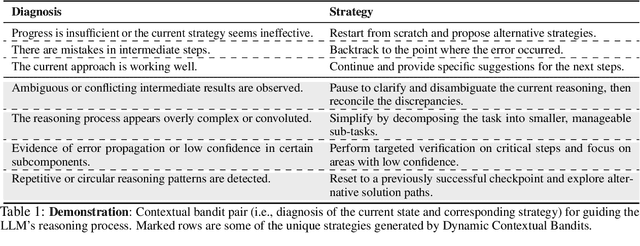
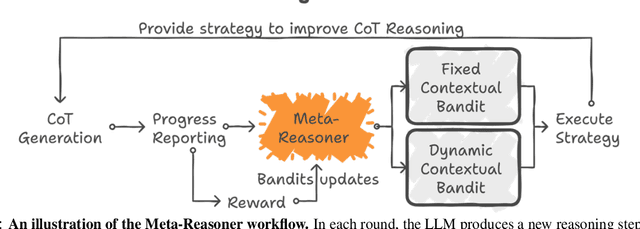
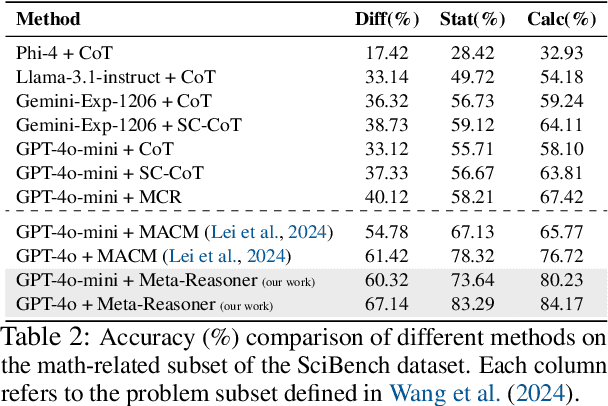
Abstract:Large Language Models (LLMs) increasingly rely on prolonged reasoning chains to solve complex tasks. However, this trial-and-error approach often leads to high computational overhead and error propagation, where early mistakes can derail subsequent steps. To address these issues, we introduce Meta-Reasoner, a framework that dynamically optimizes inference-time reasoning by enabling LLMs to "think about how to think." Drawing inspiration from human meta-cognition and dual-process theory, Meta-Reasoner operates as a strategic advisor, decoupling high-level guidance from step-by-step generation. It employs "contextual multi-armed bandits" to iteratively evaluate reasoning progress, and select optimal strategies (e.g., backtrack, clarify ambiguity, restart from scratch, or propose alternative approaches), and reallocates computational resources toward the most promising paths. Our evaluations on mathematical reasoning and puzzles highlight the potential of dynamic reasoning chains to overcome inherent challenges in the LLM reasoning process and also show promise in broader applications, offering a scalable and adaptable solution for reasoning-intensive tasks.
CovHuSeg: An Enhanced Approach for Kidney Pathology Segmentation
Nov 28, 2024



Abstract:Segmentation has long been essential in computer vision due to its numerous real-world applications. However, most traditional deep learning and machine learning models need help to capture geometric features such as size and convexity of the segmentation targets, resulting in suboptimal outcomes. To resolve this problem, we propose using a CovHuSeg algorithm to solve the problem of kidney glomeruli segmentation. This simple post-processing method is specified to adapt to the segmentation of ball-shaped anomalies, including the glomerulus. Unlike other post-processing methods, the CovHuSeg algorithm assures that the outcome mask does not have holes in it or comes in unusual shapes that are impossible to be the shape of a glomerulus. We illustrate the effectiveness of our method by experimenting with multiple deep-learning models in the context of segmentation on kidney pathology images. The results show that all models have increased accuracy when using the CovHuSeg algorithm.
Are Anomaly Scores Telling the Whole Story? A Benchmark for Multilevel Anomaly Detection
Nov 21, 2024



Abstract:Anomaly detection (AD) is a machine learning task that identifies anomalies by learning patterns from normal training data. In many real-world scenarios, anomalies vary in severity, from minor anomalies with little risk to severe abnormalities requiring immediate attention. However, existing models primarily operate in a binary setting, and the anomaly scores they produce are usually based on the deviation of data points from normal data, which may not accurately reflect practical severity. In this paper, we address this gap by making three key contributions. First, we propose a novel setting, Multilevel AD (MAD), in which the anomaly score represents the severity of anomalies in real-world applications, and we highlight its diverse applications across various domains. Second, we introduce a novel benchmark, MAD-Bench, that evaluates models not only on their ability to detect anomalies, but also on how effectively their anomaly scores reflect severity. This benchmark incorporates multiple types of baselines and real-world applications involving severity. Finally, we conduct a comprehensive performance analysis on MAD-Bench. We evaluate models on their ability to assign severity-aligned scores, investigate the correspondence between their performance on binary and multilevel detection, and study their robustness. This analysis offers key insights into improving AD models for practical severity alignment. The code framework and datasets used for the benchmark will be made publicly available.
KnowPhish: Large Language Models Meet Multimodal Knowledge Graphs for Enhancing Reference-Based Phishing Detection
Mar 04, 2024



Abstract:Phishing attacks have inflicted substantial losses on individuals and businesses alike, necessitating the development of robust and efficient automated phishing detection approaches. Reference-based phishing detectors (RBPDs), which compare the logos on a target webpage to a known set of logos, have emerged as the state-of-the-art approach. However, a major limitation of existing RBPDs is that they rely on a manually constructed brand knowledge base, making it infeasible to scale to a large number of brands, which results in false negative errors due to the insufficient brand coverage of the knowledge base. To address this issue, we propose an automated knowledge collection pipeline, using which we collect and release a large-scale multimodal brand knowledge base, KnowPhish, containing 20k brands with rich information about each brand. KnowPhish can be used to boost the performance of existing RBPDs in a plug-and-play manner. A second limitation of existing RBPDs is that they solely rely on the image modality, ignoring useful textual information present in the webpage HTML. To utilize this textual information, we propose a Large Language Model (LLM)-based approach to extract brand information of webpages from text. Our resulting multimodal phishing detection approach, KnowPhish Detector (KPD), can detect phishing webpages with or without logos. We evaluate KnowPhish and KPD on a manually validated dataset, and on a field study under Singapore's local context, showing substantial improvements in effectiveness and efficiency compared to state-of-the-art baselines.
LVM-Med: Learning Large-Scale Self-Supervised Vision Models for Medical Imaging via Second-order Graph Matching
Jul 09, 2023



Abstract:Obtaining large pre-trained models that can be fine-tuned to new tasks with limited annotated samples has remained an open challenge for medical imaging data. While pre-trained deep networks on ImageNet and vision-language foundation models trained on web-scale data are prevailing approaches, their effectiveness on medical tasks is limited due to the significant domain shift between natural and medical images. To bridge this gap, we introduce LVM-Med, the first family of deep networks trained on large-scale medical datasets. We have collected approximately 1.3 million medical images from 55 publicly available datasets, covering a large number of organs and modalities such as CT, MRI, X-ray, and Ultrasound. We benchmark several state-of-the-art self-supervised algorithms on this dataset and propose a novel self-supervised contrastive learning algorithm using a graph-matching formulation. The proposed approach makes three contributions: (i) it integrates prior pair-wise image similarity metrics based on local and global information; (ii) it captures the structural constraints of feature embeddings through a loss function constructed via a combinatorial graph-matching objective; and (iii) it can be trained efficiently end-to-end using modern gradient-estimation techniques for black-box solvers. We thoroughly evaluate the proposed LVM-Med on 15 downstream medical tasks ranging from segmentation and classification to object detection, and both for the in and out-of-distribution settings. LVM-Med empirically outperforms a number of state-of-the-art supervised, self-supervised, and foundation models. For challenging tasks such as Brain Tumor Classification or Diabetic Retinopathy Grading, LVM-Med improves previous vision-language models trained on 1 billion masks by 6-7% while using only a ResNet-50.
Anomaly Detection under Distribution Shift
Mar 24, 2023



Abstract:Anomaly detection (AD) is a crucial machine learning task that aims to learn patterns from a set of normal training samples to identify abnormal samples in test data. Most existing AD studies assume that the training and test data are drawn from the same data distribution, but the test data can have large distribution shifts arising in many real-world applications due to different natural variations such as new lighting conditions, object poses, or background appearances, rendering existing AD methods ineffective in such cases. In this paper, we consider the problem of anomaly detection under distribution shift and establish performance benchmarks on three widely-used AD and out-of-distribution (OOD) generalization datasets. We demonstrate that simple adaptation of state-of-the-art OOD generalization methods to AD settings fails to work effectively due to the lack of labeled anomaly data. We further introduce a novel robust AD approach to diverse distribution shifts by minimizing the distribution gap between in-distribution and OOD normal samples in both the training and inference stages in an unsupervised way. Our extensive empirical results on the three datasets show that our approach substantially outperforms state-of-the-art AD methods and OOD generalization methods on data with various distribution shifts, while maintaining the detection accuracy on in-distribution data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge