Armin Gruber
DomainLab: A modular Python package for domain generalization in deep learning
Mar 21, 2024
Abstract:Poor generalization performance caused by distribution shifts in unseen domains often hinders the trustworthy deployment of deep neural networks. Many domain generalization techniques address this problem by adding a domain invariant regularization loss terms during training. However, there is a lack of modular software that allows users to combine the advantages of different methods with minimal effort for reproducibility. DomainLab is a modular Python package for training user specified neural networks with composable regularization loss terms. Its decoupled design allows the separation of neural networks from regularization loss construction. Hierarchical combinations of neural networks, different domain generalization methods, and associated hyperparameters, can all be specified together with other experimental setup in a single configuration file. Hierarchical combinations of neural networks, different domain generalization methods, and associated hyperparameters, can all be specified together with other experimental setup in a single configuration file. In addition, DomainLab offers powerful benchmarking functionality to evaluate the generalization performance of neural networks in out-of-distribution data. The package supports running the specified benchmark on an HPC cluster or on a standalone machine. The package is well tested with over 95 percent coverage and well documented. From the user perspective, it is closed to modification but open to extension. The package is under the MIT license, and its source code, tutorial and documentation can be found at https://github.com/marrlab/DomainLab.
A Continual Learning Approach for Cross-Domain White Blood Cell Classification
Aug 24, 2023



Abstract:Accurate classification of white blood cells in peripheral blood is essential for diagnosing hematological diseases. Due to constantly evolving clinical settings, data sources, and disease classifications, it is necessary to update machine learning classification models regularly for practical real-world use. Such models significantly benefit from sequentially learning from incoming data streams without forgetting previously acquired knowledge. However, models can suffer from catastrophic forgetting, causing a drop in performance on previous tasks when fine-tuned on new data. Here, we propose a rehearsal-based continual learning approach for class incremental and domain incremental scenarios in white blood cell classification. To choose representative samples from previous tasks, we employ exemplar set selection based on the model's predictions. This involves selecting the most confident samples and the most challenging samples identified through uncertainty estimation of the model. We thoroughly evaluated our proposed approach on three white blood cell classification datasets that differ in color, resolution, and class composition, including scenarios where new domains or new classes are introduced to the model with every task. We also test a long class incremental experiment with both new domains and new classes. Our results demonstrate that our approach outperforms established baselines in continual learning, including existing iCaRL and EWC methods for classifying white blood cells in cross-domain environments.
Imbalanced Domain Generalization for Robust Single Cell Classification in Hematological Cytomorphology
Mar 14, 2023



Abstract:Accurate morphological classification of white blood cells (WBCs) is an important step in the diagnosis of leukemia, a disease in which nonfunctional blast cells accumulate in the bone marrow. Recently, deep convolutional neural networks (CNNs) have been successfully used to classify leukocytes by training them on single-cell images from a specific domain. Most CNN models assume that the distributions of the training and test data are similar, i.e., that the data are independently and identically distributed. Therefore, they are not robust to different staining protocols, magnifications, resolutions, scanners, or imaging protocols, as well as variations in clinical centers or patient cohorts. In addition, domain-specific data imbalances affect the generalization performance of classifiers. Here, we train a robust CNN for WBC classification by addressing cross-domain data imbalance and domain shifts. To this end, we use two loss functions and demonstrate the effectiveness on out-of-distribution (OOD) generalization. Our approach achieves the best F1 macro score compared to other existing methods, and is able to consider rare cell types. This is the first demonstration of imbalanced domain generalization in hematological cytomorphology and paves the way for robust single cell classification methods for the application in laboratories and clinics.
Unsupervised Cross-Domain Feature Extraction for Single Blood Cell Image Classification
Jul 01, 2022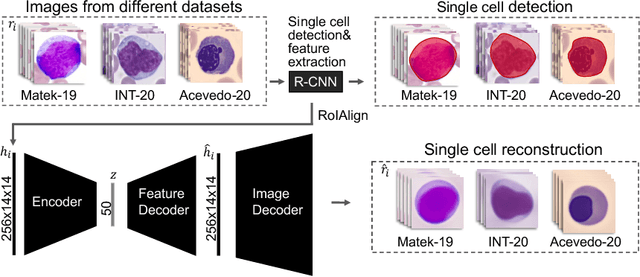
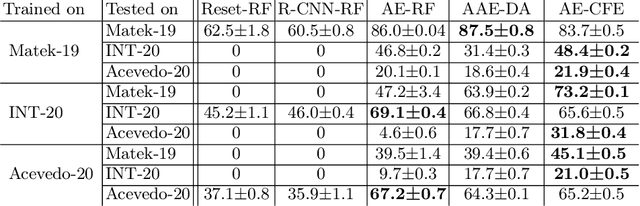
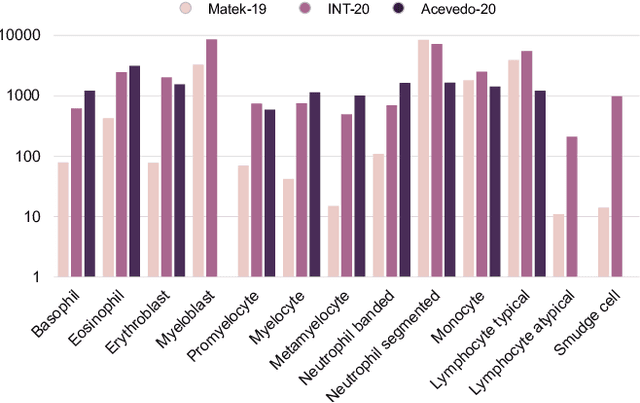
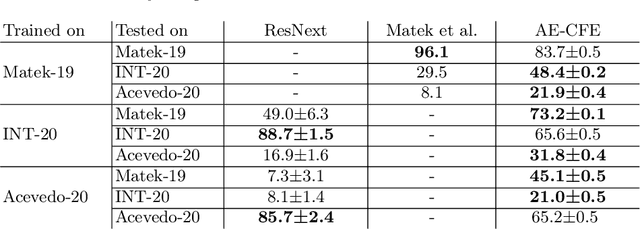
Abstract:Diagnosing hematological malignancies requires identification and classification of white blood cells in peripheral blood smears. Domain shifts caused by different lab procedures, staining, illumination, and microscope settings hamper the re-usability of recently developed machine learning methods on data collected from different sites. Here, we propose a cross-domain adapted autoencoder to extract features in an unsupervised manner on three different datasets of single white blood cells scanned from peripheral blood smears. The autoencoder is based on an R-CNN architecture allowing it to focus on the relevant white blood cell and eliminate artifacts in the image. To evaluate the quality of the extracted features we use a simple random forest to classify single cells. We show that thanks to the rich features extracted by the autoencoder trained on only one of the datasets, the random forest classifier performs satisfactorily on the unseen datasets, and outperforms published oracle networks in the cross-domain task. Our results suggest the possibility of employing this unsupervised approach in more complicated diagnosis and prognosis tasks without the need to add expensive expert labels to unseen data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge