Patrick Rockenschaub
Robust prediction under missingness shifts
Jun 24, 2024Abstract:Prediction becomes more challenging with missing covariates. What method is chosen to handle missingness can greatly affect how models perform. In many real-world problems, the best prediction performance is achieved by models that can leverage the informative nature of a value being missing. Yet, the reasons why a covariate goes missing can change once a model is deployed in practice. If such a missingness shift occurs, the conditional probability of a value being missing differs in the target data. Prediction performance in the source data may no longer be a good selection criterion, and approaches that do not rely on informative missingness may be preferable. However, we show that the Bayes predictor remains unchanged by ignorable shifts for which the probability of missingness only depends on observed data. Any consistent estimator of the Bayes predictor may therefore result in robust prediction under those conditions, although we show empirically that different methods appear robust to different types of shifts. If the missingness shift is non-ignorable, the Bayes predictor may change due to the shift. While neither approach recovers the Bayes predictor in this case, we found empirically that disregarding missingness was most beneficial when it was highly informative.
DomainLab: A modular Python package for domain generalization in deep learning
Mar 21, 2024
Abstract:Poor generalization performance caused by distribution shifts in unseen domains often hinders the trustworthy deployment of deep neural networks. Many domain generalization techniques address this problem by adding a domain invariant regularization loss terms during training. However, there is a lack of modular software that allows users to combine the advantages of different methods with minimal effort for reproducibility. DomainLab is a modular Python package for training user specified neural networks with composable regularization loss terms. Its decoupled design allows the separation of neural networks from regularization loss construction. Hierarchical combinations of neural networks, different domain generalization methods, and associated hyperparameters, can all be specified together with other experimental setup in a single configuration file. Hierarchical combinations of neural networks, different domain generalization methods, and associated hyperparameters, can all be specified together with other experimental setup in a single configuration file. In addition, DomainLab offers powerful benchmarking functionality to evaluate the generalization performance of neural networks in out-of-distribution data. The package supports running the specified benchmark on an HPC cluster or on a standalone machine. The package is well tested with over 95 percent coverage and well documented. From the user perspective, it is closed to modification but open to extension. The package is under the MIT license, and its source code, tutorial and documentation can be found at https://github.com/marrlab/DomainLab.
Yet Another ICU Benchmark: A Flexible Multi-Center Framework for Clinical ML
Jun 08, 2023Abstract:Medical applications of machine learning (ML) have experienced a surge in popularity in recent years. The intensive care unit (ICU) is a natural habitat for ML given the abundance of available data from electronic health records. Models have been proposed to address numerous ICU prediction tasks like the early detection of complications. While authors frequently report state-of-the-art performance, it is challenging to verify claims of superiority. Datasets and code are not always published, and cohort definitions, preprocessing pipelines, and training setups are difficult to reproduce. This work introduces Yet Another ICU Benchmark (YAIB), a modular framework that allows researchers to define reproducible and comparable clinical ML experiments; we offer an end-to-end solution from cohort definition to model evaluation. The framework natively supports most open-access ICU datasets (MIMIC III/IV, eICU, HiRID, AUMCdb) and is easily adaptable to future ICU datasets. Combined with a transparent preprocessing pipeline and extensible training code for multiple ML and deep learning models, YAIB enables unified model development. Our benchmark comes with five predefined established prediction tasks (mortality, acute kidney injury, sepsis, kidney function, and length of stay) developed in collaboration with clinicians. Adding further tasks is straightforward by design. Using YAIB, we demonstrate that the choice of dataset, cohort definition, and preprocessing have a major impact on the prediction performance - often more so than model class - indicating an urgent need for YAIB as a holistic benchmarking tool. We provide our work to the clinical ML community to accelerate method development and enable real-world clinical implementations. Software Repository: https://github.com/rvandewater/YAIB.
From Single-Hospital to Multi-Centre Applications: Enhancing the Generalisability of Deep Learning Models for Adverse Event Prediction in the ICU
Apr 07, 2023



Abstract:Deep learning (DL) can aid doctors in detecting worsening patient states early, affording them time to react and prevent bad outcomes. While DL-based early warning models usually work well in the hospitals they were trained for, they tend to be less reliable when applied at new hospitals. This makes it difficult to deploy them at scale. Using carefully harmonised intensive care data from four data sources across Europe and the US (totalling 334,812 stays), we systematically assessed the reliability of DL models for three common adverse events: death, acute kidney injury (AKI), and sepsis. We tested whether using more than one data source and/or explicitly optimising for generalisability during training improves model performance at new hospitals. We found that models achieved high AUROC for mortality (0.838-0.869), AKI (0.823-0.866), and sepsis (0.749-0.824) at the training hospital. As expected, performance dropped at new hospitals, sometimes by as much as -0.200. Using more than one data source for training mitigated the performance drop, with multi-source models performing roughly on par with the best single-source model. This suggests that as data from more hospitals become available for training, model robustness is likely to increase, lower-bounding robustness with the performance of the most applicable data source in the training data. Dedicated methods promoting generalisability did not noticeably improve performance in our experiments.
Longitudinal patient stratification of electronic health records with flexible adjustment for clinical outcomes
Nov 11, 2021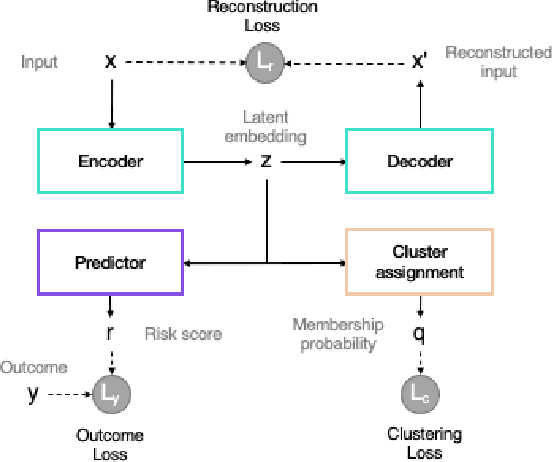
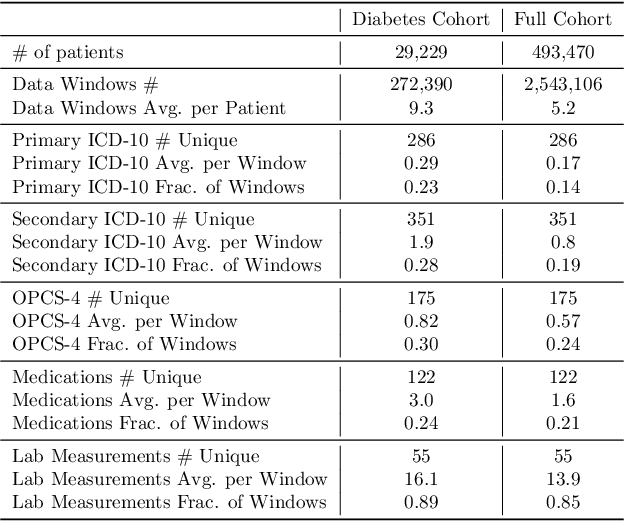
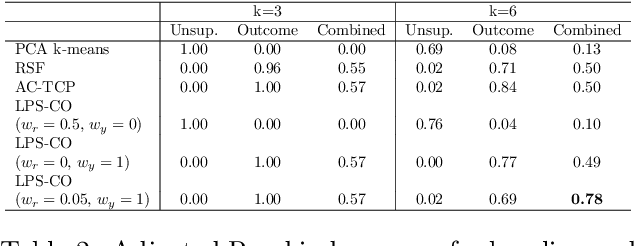
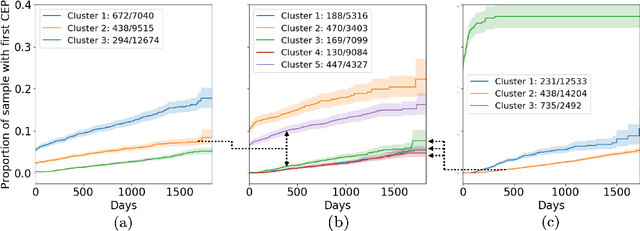
Abstract:The increase in availability of longitudinal electronic health record (EHR) data is leading to improved understanding of diseases and discovery of novel phenotypes. The majority of clustering algorithms focus only on patient trajectories, yet patients with similar trajectories may have different outcomes. Finding subgroups of patients with different trajectories and outcomes can guide future drug development and improve recruitment to clinical trials. We develop a recurrent neural network autoencoder to cluster EHR data using reconstruction, outcome, and clustering losses which can be weighted to find different types of patient clusters. We show our model is able to discover known clusters from both data biases and outcome differences, outperforming baseline models. We demonstrate the model performance on $29,229$ diabetes patients, showing it finds clusters of patients with both different trajectories and different outcomes which can be utilized to aid clinical decision making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge