Owen Parsons
Sensyne Health, Oxford, UK
Enabling scalable clinical interpretation of ML-based phenotypes using real world data
Aug 02, 2022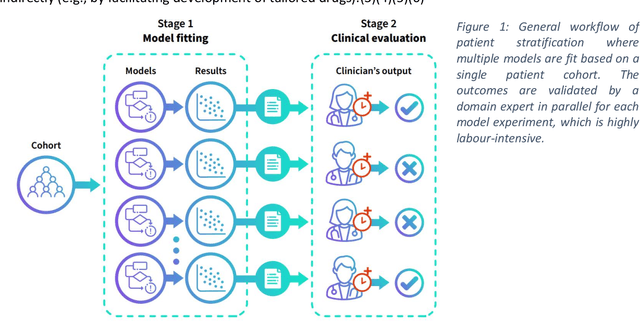
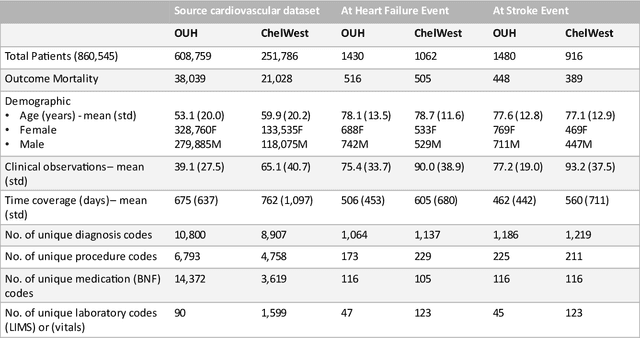
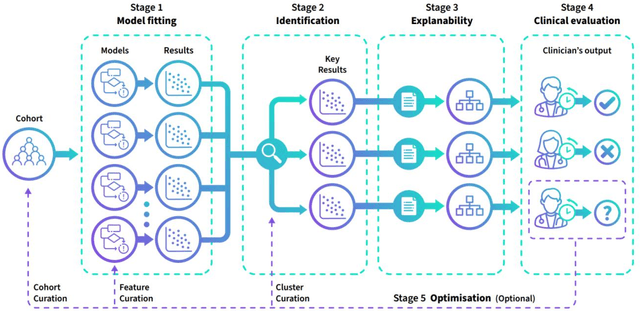
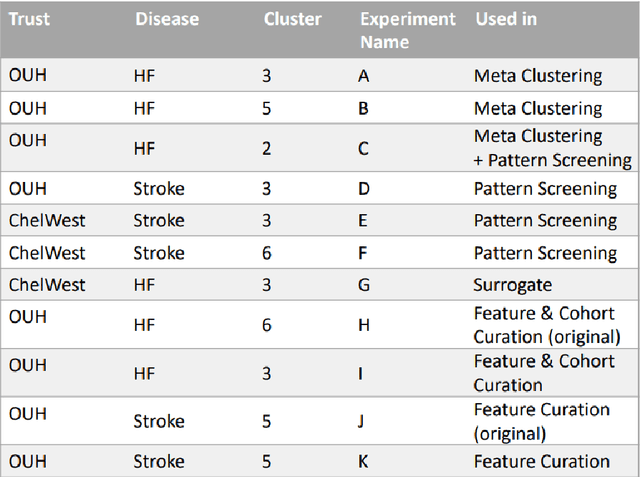
Abstract:The availability of large and deep electronic healthcare records (EHR) datasets has the potential to enable a better understanding of real-world patient journeys, and to identify novel subgroups of patients. ML-based aggregation of EHR data is mostly tool-driven, i.e., building on available or newly developed methods. However, these methods, their input requirements, and, importantly, resulting output are frequently difficult to interpret, especially without in-depth data science or statistical training. This endangers the final step of analysis where an actionable and clinically meaningful interpretation is needed.This study investigates approaches to perform patient stratification analysis at scale using large EHR datasets and multiple clustering methods for clinical research. We have developed several tools to facilitate the clinical evaluation and interpretation of unsupervised patient stratification results, namely pattern screening, meta clustering, surrogate modeling, and curation. These tools can be used at different stages within the analysis. As compared to a standard analysis approach, we demonstrate the ability to condense results and optimize analysis time. In the case of meta clustering, we demonstrate that the number of patient clusters can be reduced from 72 to 3 in one example. In another stratification result, by using surrogate models, we could quickly identify that heart failure patients were stratified if blood sodium measurements were available. As this is a routine measurement performed for all patients with heart failure, this indicated a data bias. By using further cohort and feature curation, these patients and other irrelevant features could be removed to increase the clinical meaningfulness. These examples show the effectiveness of the proposed methods and we hope to encourage further research in this field.
Compensating trajectory bias for unsupervised patient stratification using adversarial recurrent neural networks
Dec 14, 2021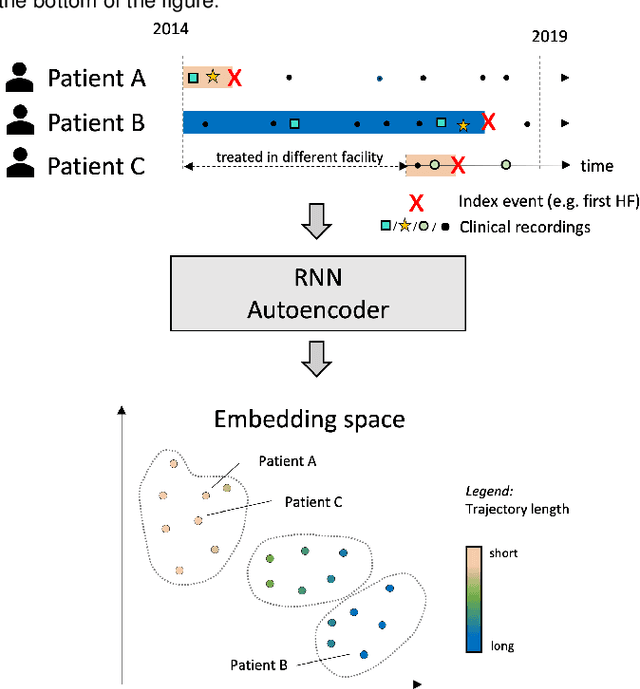
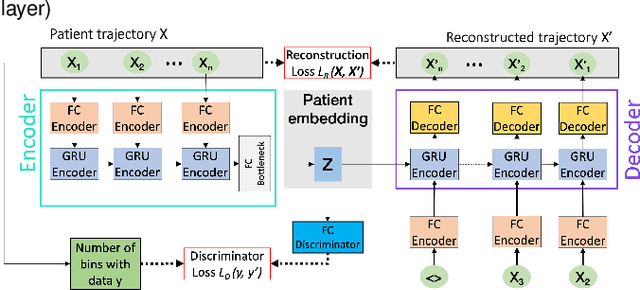
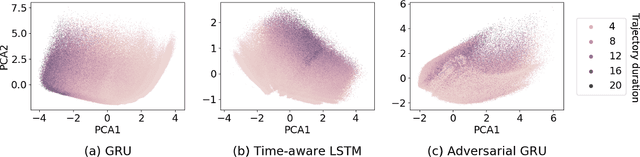
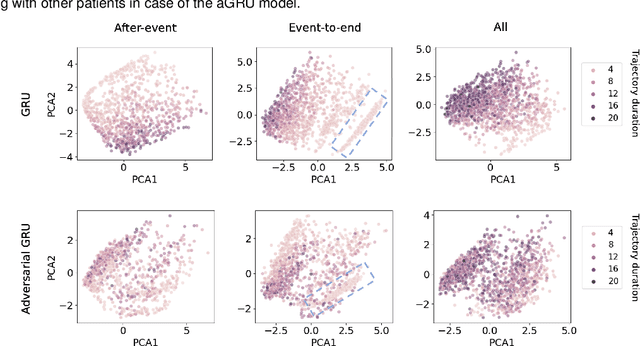
Abstract:Electronic healthcare records are an important source of information which can be used in patient stratification to discover novel disease phenotypes. However, they can be challenging to work with as data is often sparse and irregularly sampled. One approach to solve these limitations is learning dense embeddings that represent individual patient trajectories using a recurrent neural network autoencoder (RNN-AE). This process can be susceptible to unwanted data biases. We show that patient embeddings and clusters using previously proposed RNN-AE models might be impacted by a trajectory bias, meaning that results are dominated by the amount of data contained in each patients trajectory, instead of clinically relevant details. We investigate this bias on 2 datasets (from different hospitals) and 2 disease areas as well as using different parts of the patient trajectory. Our results using 2 previously published baseline methods indicate a particularly strong bias in case of an event-to-end trajectory. We present a method that can overcome this issue using an adversarial training scheme on top of a RNN-AE. Our results show that our approach can reduce the trajectory bias in all cases.
Longitudinal patient stratification of electronic health records with flexible adjustment for clinical outcomes
Nov 11, 2021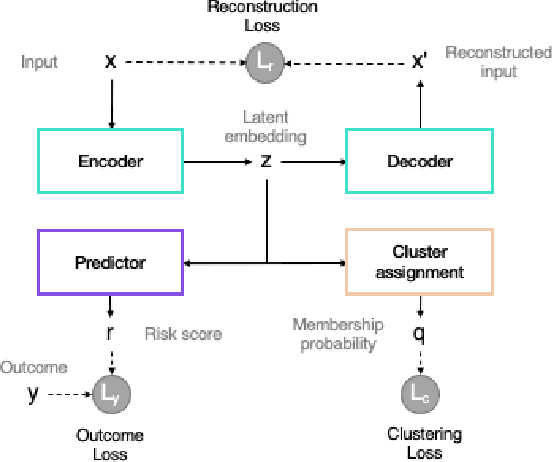
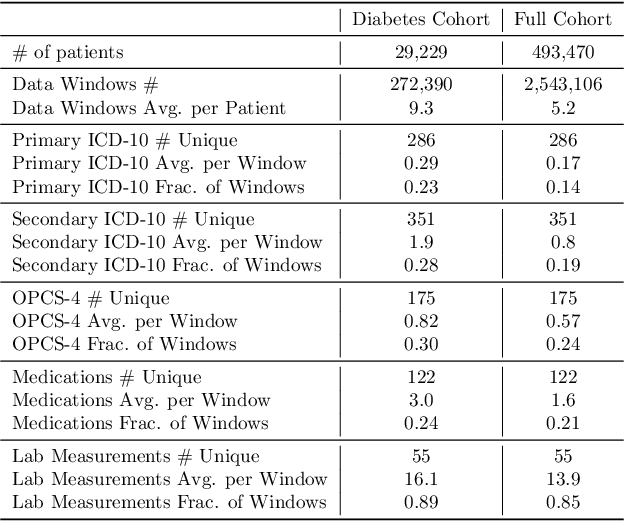
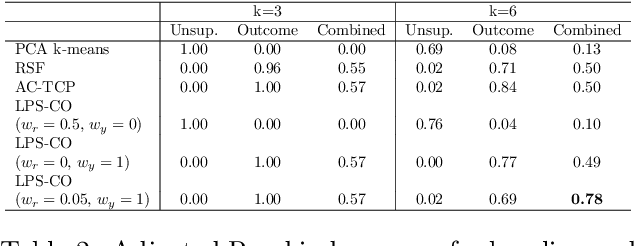
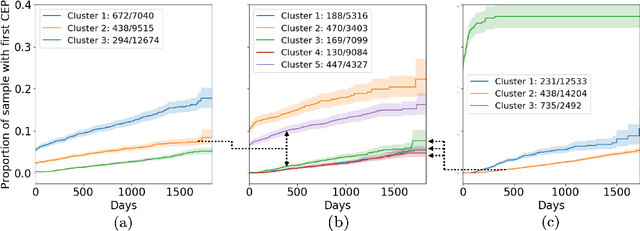
Abstract:The increase in availability of longitudinal electronic health record (EHR) data is leading to improved understanding of diseases and discovery of novel phenotypes. The majority of clustering algorithms focus only on patient trajectories, yet patients with similar trajectories may have different outcomes. Finding subgroups of patients with different trajectories and outcomes can guide future drug development and improve recruitment to clinical trials. We develop a recurrent neural network autoencoder to cluster EHR data using reconstruction, outcome, and clustering losses which can be weighted to find different types of patient clusters. We show our model is able to discover known clusters from both data biases and outcome differences, outperforming baseline models. We demonstrate the model performance on $29,229$ diabetes patients, showing it finds clusters of patients with both different trajectories and different outcomes which can be utilized to aid clinical decision making.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge