Robert Dürichen
Sensyne Health, Oxford, UK
Enabling scalable clinical interpretation of ML-based phenotypes using real world data
Aug 02, 2022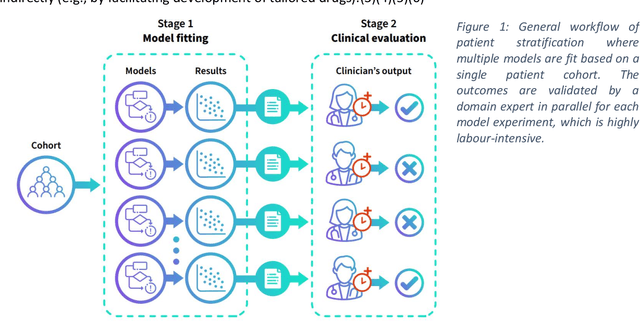
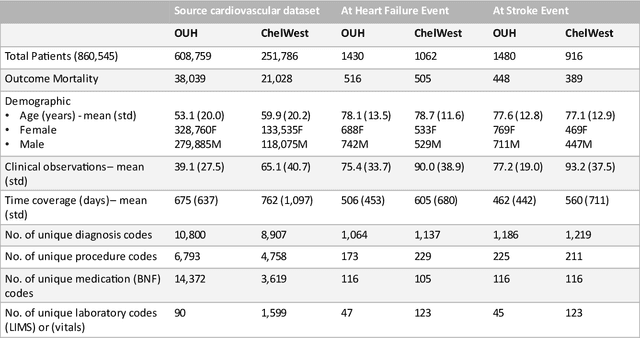
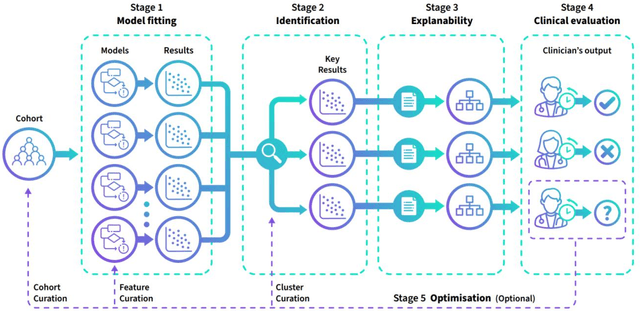
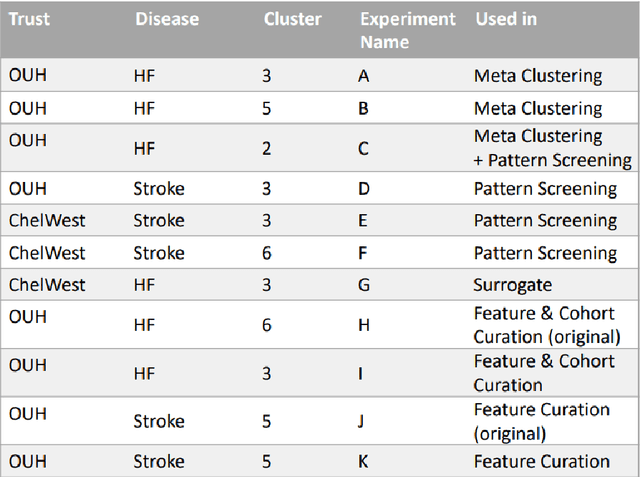
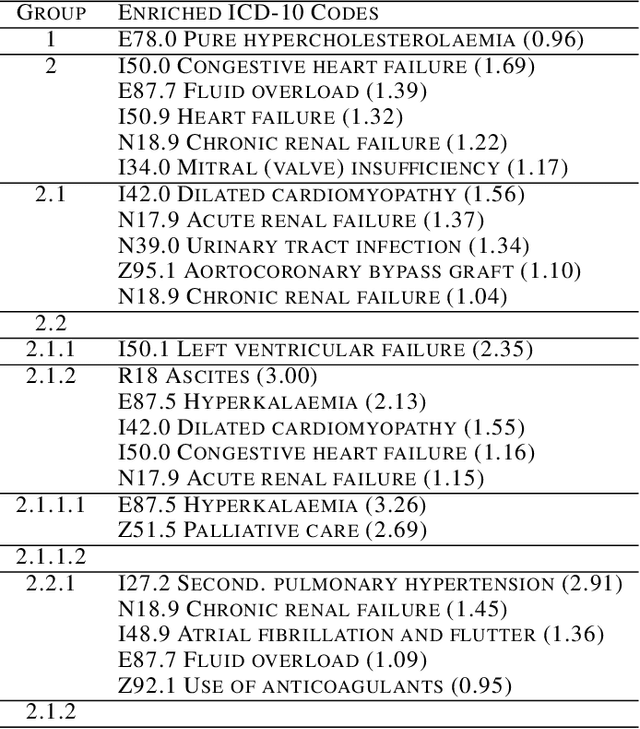
Abstract:The availability of large and deep electronic healthcare records (EHR) datasets has the potential to enable a better understanding of real-world patient journeys, and to identify novel subgroups of patients. ML-based aggregation of EHR data is mostly tool-driven, i.e., building on available or newly developed methods. However, these methods, their input requirements, and, importantly, resulting output are frequently difficult to interpret, especially without in-depth data science or statistical training. This endangers the final step of analysis where an actionable and clinically meaningful interpretation is needed.This study investigates approaches to perform patient stratification analysis at scale using large EHR datasets and multiple clustering methods for clinical research. We have developed several tools to facilitate the clinical evaluation and interpretation of unsupervised patient stratification results, namely pattern screening, meta clustering, surrogate modeling, and curation. These tools can be used at different stages within the analysis. As compared to a standard analysis approach, we demonstrate the ability to condense results and optimize analysis time. In the case of meta clustering, we demonstrate that the number of patient clusters can be reduced from 72 to 3 in one example. In another stratification result, by using surrogate models, we could quickly identify that heart failure patients were stratified if blood sodium measurements were available. As this is a routine measurement performed for all patients with heart failure, this indicated a data bias. By using further cohort and feature curation, these patients and other irrelevant features could be removed to increase the clinical meaningfulness. These examples show the effectiveness of the proposed methods and we hope to encourage further research in this field.
Similarity-based prediction of Ejection Fraction in Heart Failure Patients
Mar 14, 2022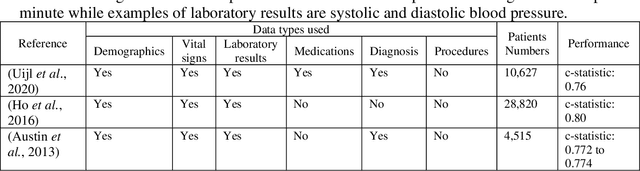
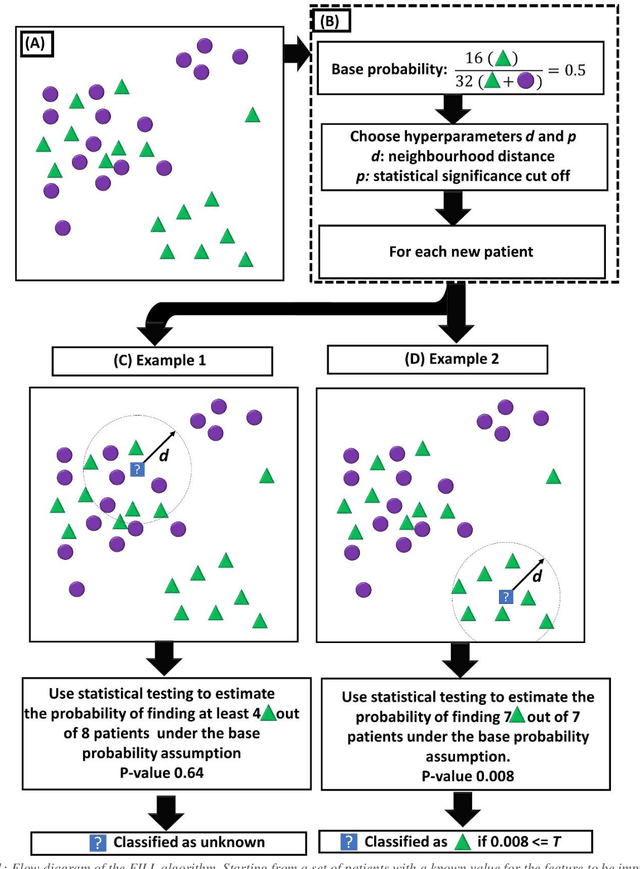


Abstract:Biomedical research is increasingly employing real world evidence (RWE) to foster discoveries of novel clinical phenotypes and to better characterize long term effect of medical treatments. However, due to limitations inherent in the collection process, RWE often lacks key features of patients, particularly when these features cannot be directly encoded using data standards such as ICD-10. Here we propose a novel data-driven statistical machine learning approach, named Feature Imputation via Local Likelihood (FILL), designed to infer missing features by exploiting feature similarity between patients. We test our method using a particularly challenging problem: differentiating heart failure patients with reduced versus preserved ejection fraction (HFrEF and HFpEF respectively). The complexity of the task stems from three aspects: the two share many common characteristics and treatments, only part of the relevant diagnoses may have been recorded, and the information on ejection fraction is often missing from RWE datasets. Despite these difficulties, our method is shown to be capable of inferring heart failure patients with HFpEF with a precision above 80% when considering multiple scenarios across two RWE datasets containing 11,950 and 10,051 heart failure patients. This is an improvement when compared to classical approaches such as logistic regression and random forest which were only able to achieve a precision < 73%. Finally, this approach allows us to analyse which features are commonly associated with HFpEF patients. For example, we found that specific diagnostic codes for atrial fibrillation and personal history of long-term use of anticoagulants are often key in identifying HFpEF patients.
Compensating trajectory bias for unsupervised patient stratification using adversarial recurrent neural networks
Dec 14, 2021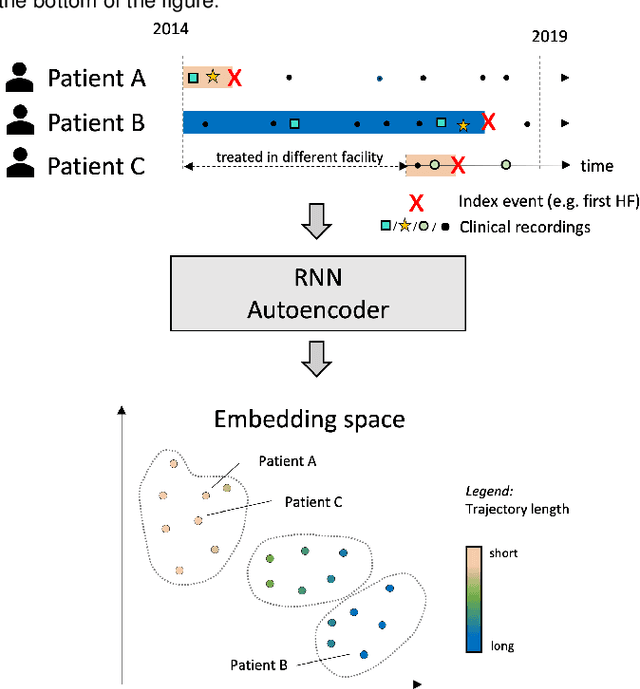
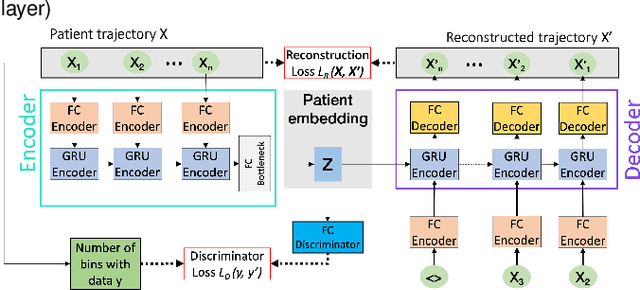
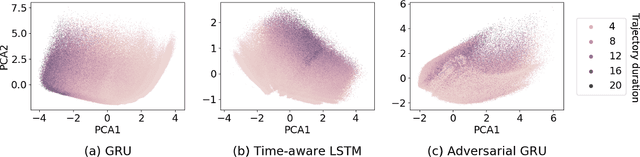
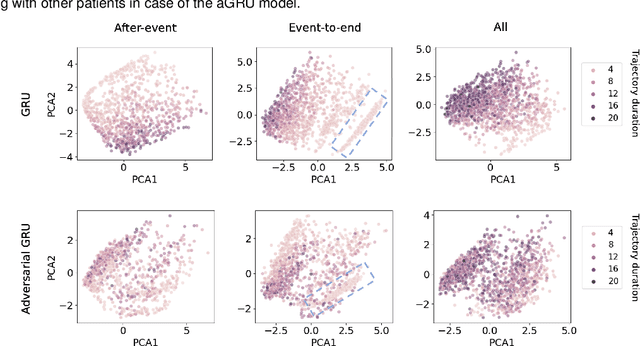
Abstract:Electronic healthcare records are an important source of information which can be used in patient stratification to discover novel disease phenotypes. However, they can be challenging to work with as data is often sparse and irregularly sampled. One approach to solve these limitations is learning dense embeddings that represent individual patient trajectories using a recurrent neural network autoencoder (RNN-AE). This process can be susceptible to unwanted data biases. We show that patient embeddings and clusters using previously proposed RNN-AE models might be impacted by a trajectory bias, meaning that results are dominated by the amount of data contained in each patients trajectory, instead of clinically relevant details. We investigate this bias on 2 datasets (from different hospitals) and 2 disease areas as well as using different parts of the patient trajectory. Our results using 2 previously published baseline methods indicate a particularly strong bias in case of an event-to-end trajectory. We present a method that can overcome this issue using an adversarial training scheme on top of a RNN-AE. Our results show that our approach can reduce the trajectory bias in all cases.
Longitudinal patient stratification of electronic health records with flexible adjustment for clinical outcomes
Nov 11, 2021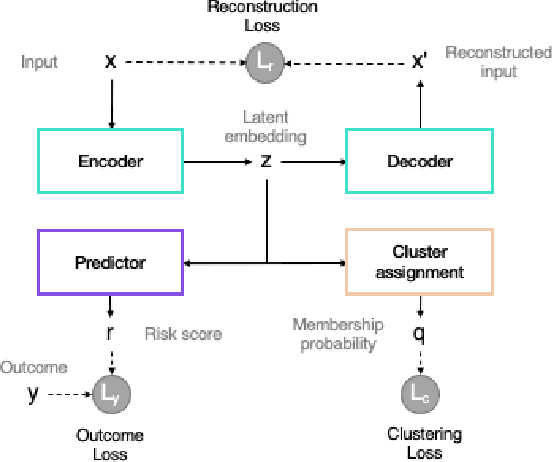
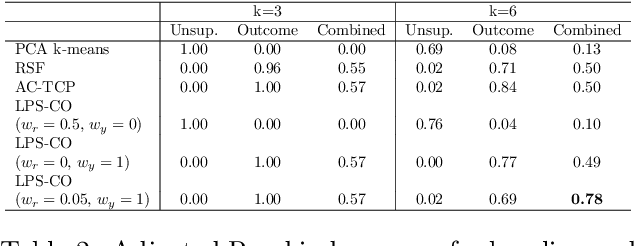
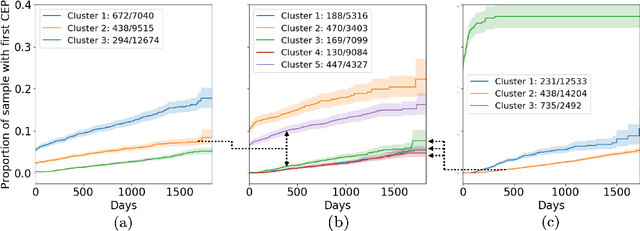
Abstract:The increase in availability of longitudinal electronic health record (EHR) data is leading to improved understanding of diseases and discovery of novel phenotypes. The majority of clustering algorithms focus only on patient trajectories, yet patients with similar trajectories may have different outcomes. Finding subgroups of patients with different trajectories and outcomes can guide future drug development and improve recruitment to clinical trials. We develop a recurrent neural network autoencoder to cluster EHR data using reconstruction, outcome, and clustering losses which can be weighted to find different types of patient clusters. We show our model is able to discover known clusters from both data biases and outcome differences, outperforming baseline models. We demonstrate the model performance on $29,229$ diabetes patients, showing it finds clusters of patients with both different trajectories and different outcomes which can be utilized to aid clinical decision making.
Deep Semi-Supervised Embedded Clustering (DSEC) for Stratification of Heart Failure Patients
Jan 17, 2021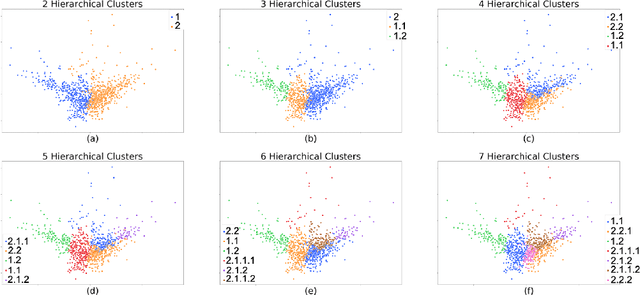
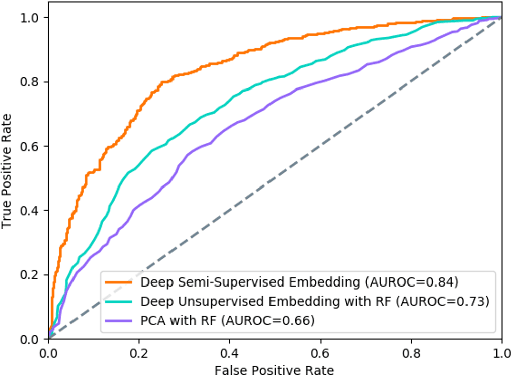
Abstract:Determining phenotypes of diseases can have considerable benefits for in-hospital patient care and to drug development. The structure of high dimensional data sets such as electronic health records are often represented through an embedding of the data, with clustering methods used to group data of similar structure. If subgroups are known to exist within data, supervised methods may be used to influence the clusters discovered. We propose to extend deep embedded clustering to a semi-supervised deep embedded clustering algorithm to stratify subgroups through known labels in the data. In this work we apply deep semi-supervised embedded clustering to determine data-driven patient subgroups of heart failure from the electronic health records of 4,487 heart failure and control patients. We find clinically relevant clusters from an embedded space derived from heterogeneous data. The proposed algorithm can potentially find new undiagnosed subgroups of patients that have different outcomes, and, therefore, lead to improved treatments.
Prediction of the onset of cardiovascular diseases from electronic health records using multi-task gated recurrent units
Jul 16, 2020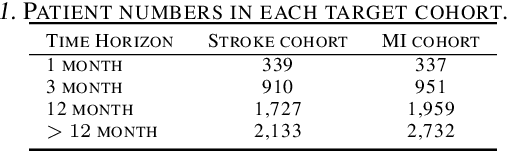
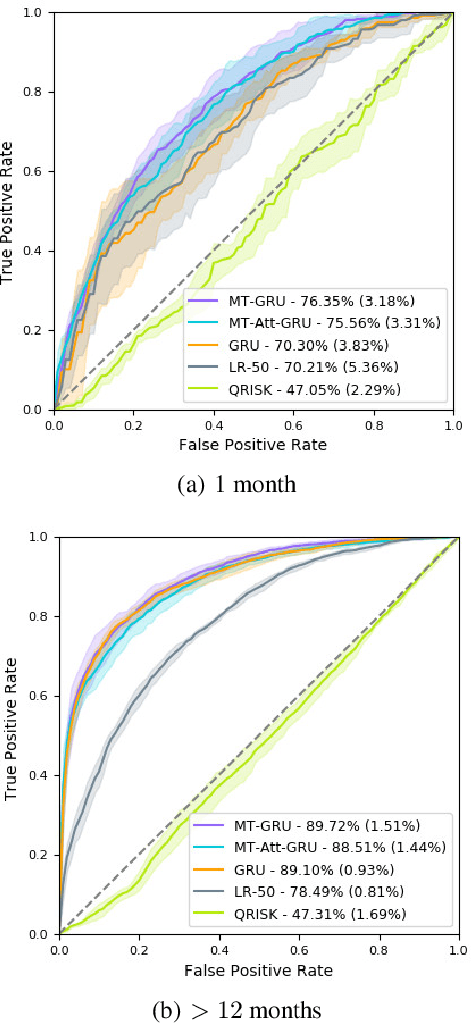
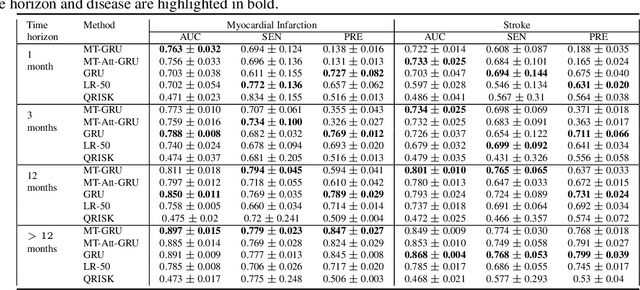
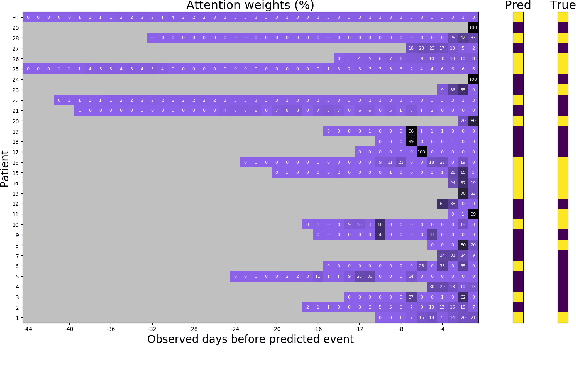
Abstract:In this work, we propose a multi-task recurrent neural network with attention mechanism for predicting cardiovascular events from electronic health records (EHRs) at different time horizons. The proposed approach is compared to a standard clinical risk predictor (QRISK) and machine learning alternatives using 5-year data from a NHS Foundation Trust. The proposed model outperforms standard clinical risk scores in predicting stroke (AUC=0.85) and myocardial infarction (AUC=0.89), considering the largest time horizon. Benefit of using an \gls{mt} setting becomes visible for very short time horizons, which results in an AUC increase between 2-6%. Further, we explored the importance of individual features and attention weights in predicting cardiovascular events. Our results indicate that the recurrent neural network approach benefits from the hospital longitudinal information and demonstrates how machine learning techniques can be applied to secondary care.
Binary Input Layer: Training of CNN models with binary input data
Dec 09, 2018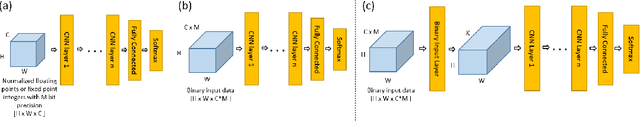
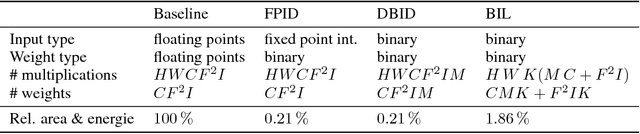
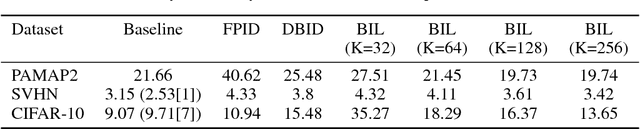
Abstract:For the efficient execution of deep convolutional neural networks (CNN) on edge devices, various approaches have been presented which reduce the bit width of the network parameters down to 1 bit. Binarization of the first layer was always excluded, as it leads to a significant error increase. Here, we present the novel concept of binary input layer (BIL), which allows the usage of binary input data by learning bit specific binary weights. The concept is evaluated on three datasets (PAMAP2, SVHN, CIFAR-10). Our results show that this approach is in particular beneficial for multimodal datasets (PAMAP2) where it outperforms networks using full precision weights in the first layer by 1:92 percentage points (pp) while consuming only 2 % of the chip area.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge