Janie Baxter
Sensyne Health, Oxford, UK
Enabling scalable clinical interpretation of ML-based phenotypes using real world data
Aug 02, 2022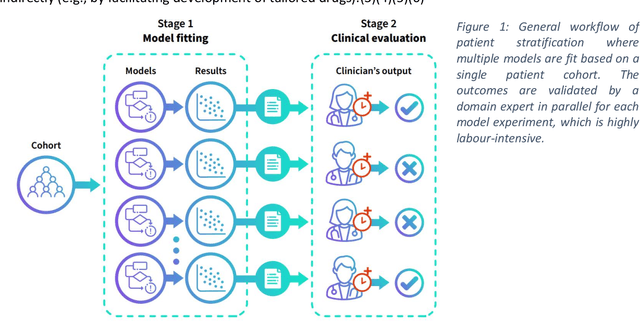
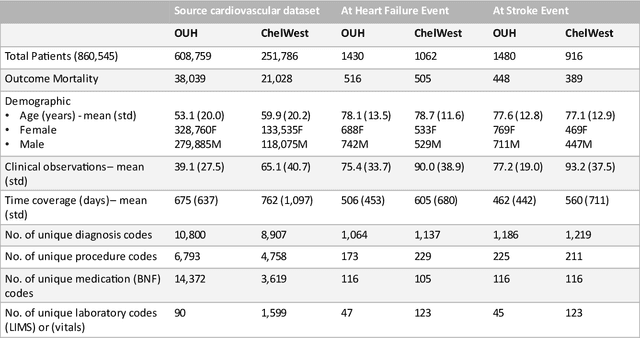
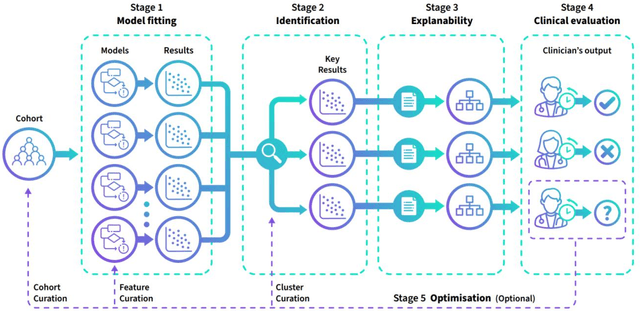
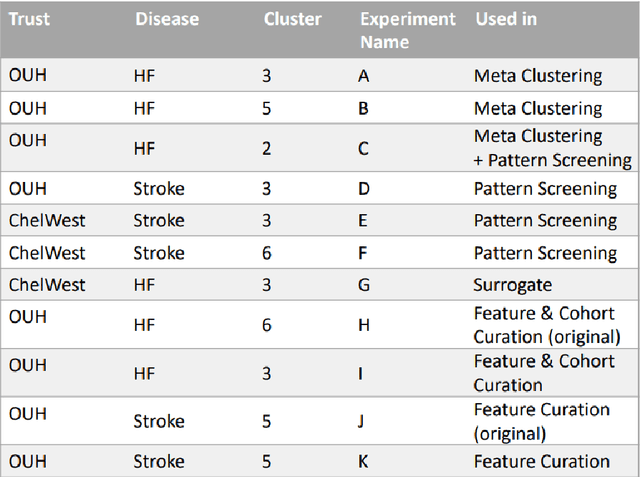
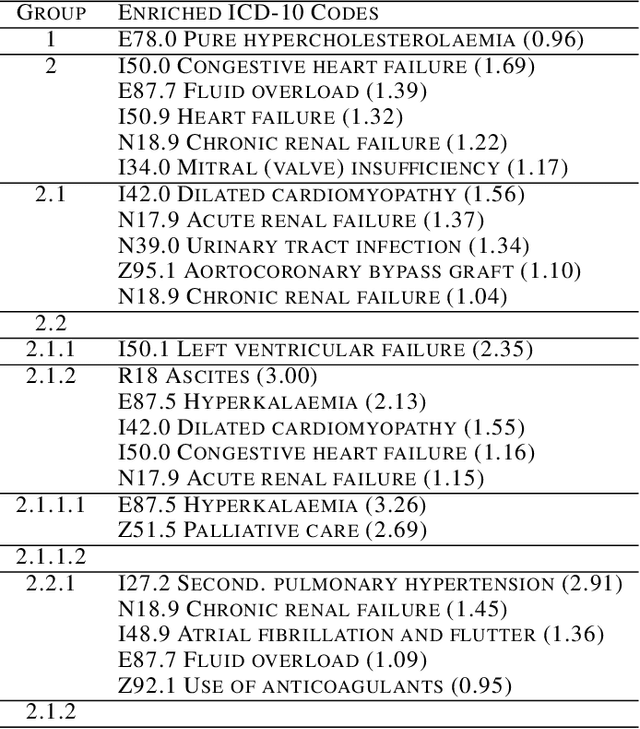
Abstract:The availability of large and deep electronic healthcare records (EHR) datasets has the potential to enable a better understanding of real-world patient journeys, and to identify novel subgroups of patients. ML-based aggregation of EHR data is mostly tool-driven, i.e., building on available or newly developed methods. However, these methods, their input requirements, and, importantly, resulting output are frequently difficult to interpret, especially without in-depth data science or statistical training. This endangers the final step of analysis where an actionable and clinically meaningful interpretation is needed.This study investigates approaches to perform patient stratification analysis at scale using large EHR datasets and multiple clustering methods for clinical research. We have developed several tools to facilitate the clinical evaluation and interpretation of unsupervised patient stratification results, namely pattern screening, meta clustering, surrogate modeling, and curation. These tools can be used at different stages within the analysis. As compared to a standard analysis approach, we demonstrate the ability to condense results and optimize analysis time. In the case of meta clustering, we demonstrate that the number of patient clusters can be reduced from 72 to 3 in one example. In another stratification result, by using surrogate models, we could quickly identify that heart failure patients were stratified if blood sodium measurements were available. As this is a routine measurement performed for all patients with heart failure, this indicated a data bias. By using further cohort and feature curation, these patients and other irrelevant features could be removed to increase the clinical meaningfulness. These examples show the effectiveness of the proposed methods and we hope to encourage further research in this field.
Compensating trajectory bias for unsupervised patient stratification using adversarial recurrent neural networks
Dec 14, 2021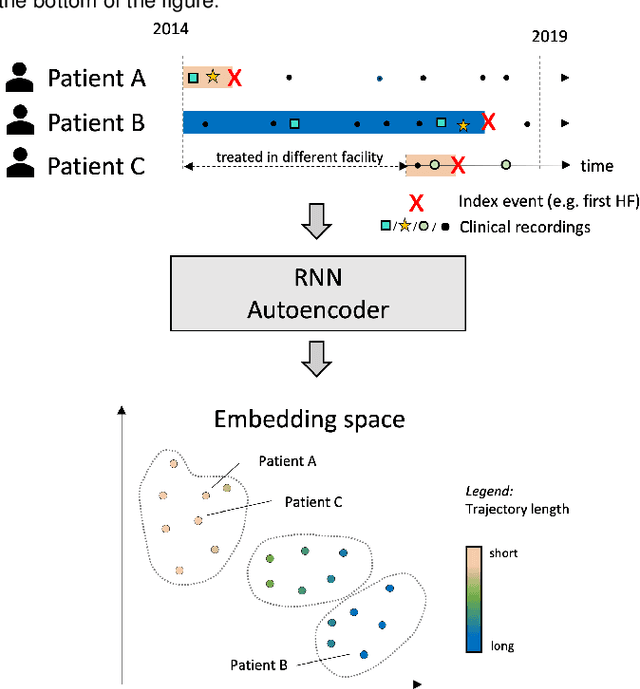
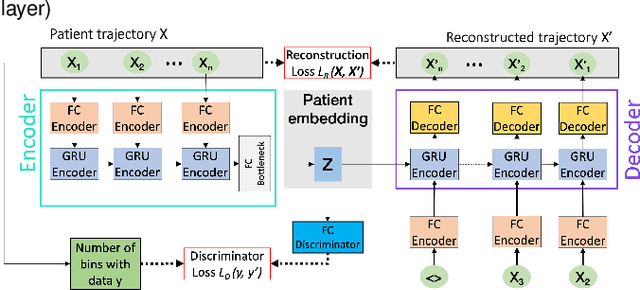
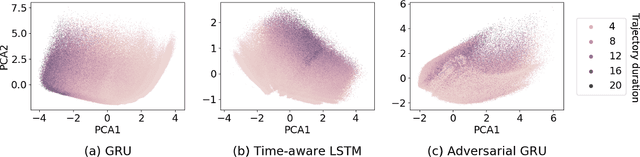
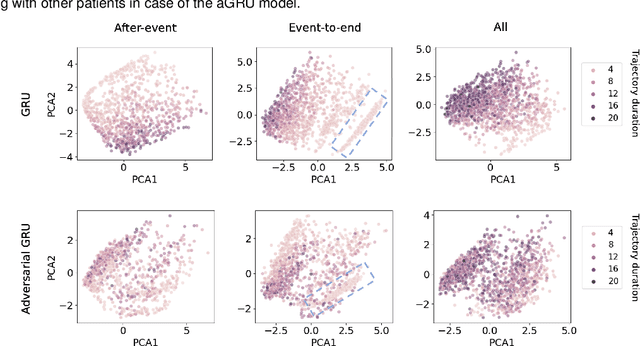
Abstract:Electronic healthcare records are an important source of information which can be used in patient stratification to discover novel disease phenotypes. However, they can be challenging to work with as data is often sparse and irregularly sampled. One approach to solve these limitations is learning dense embeddings that represent individual patient trajectories using a recurrent neural network autoencoder (RNN-AE). This process can be susceptible to unwanted data biases. We show that patient embeddings and clusters using previously proposed RNN-AE models might be impacted by a trajectory bias, meaning that results are dominated by the amount of data contained in each patients trajectory, instead of clinically relevant details. We investigate this bias on 2 datasets (from different hospitals) and 2 disease areas as well as using different parts of the patient trajectory. Our results using 2 previously published baseline methods indicate a particularly strong bias in case of an event-to-end trajectory. We present a method that can overcome this issue using an adversarial training scheme on top of a RNN-AE. Our results show that our approach can reduce the trajectory bias in all cases.
Deep Semi-Supervised Embedded Clustering (DSEC) for Stratification of Heart Failure Patients
Jan 17, 2021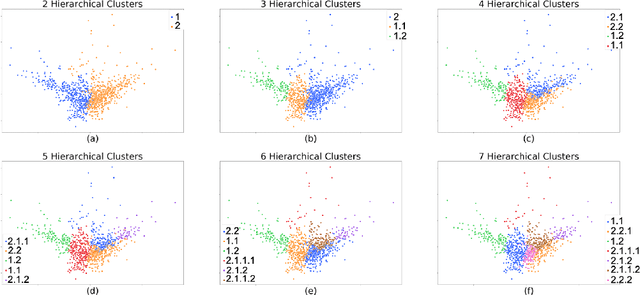
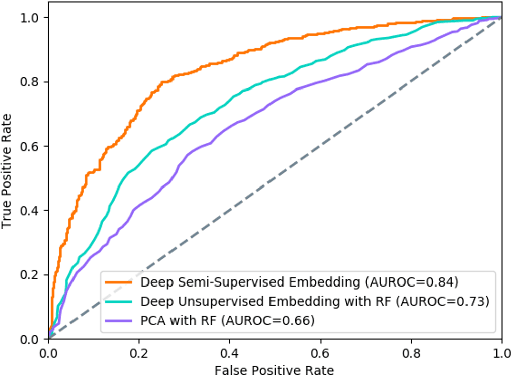
Abstract:Determining phenotypes of diseases can have considerable benefits for in-hospital patient care and to drug development. The structure of high dimensional data sets such as electronic health records are often represented through an embedding of the data, with clustering methods used to group data of similar structure. If subgroups are known to exist within data, supervised methods may be used to influence the clusters discovered. We propose to extend deep embedded clustering to a semi-supervised deep embedded clustering algorithm to stratify subgroups through known labels in the data. In this work we apply deep semi-supervised embedded clustering to determine data-driven patient subgroups of heart failure from the electronic health records of 4,487 heart failure and control patients. We find clinically relevant clusters from an embedded space derived from heterogeneous data. The proposed algorithm can potentially find new undiagnosed subgroups of patients that have different outcomes, and, therefore, lead to improved treatments.
A comparative study of artificial intelligence and human doctors for the purpose of triage and diagnosis
Jun 27, 2018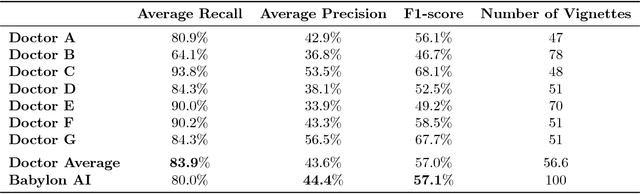
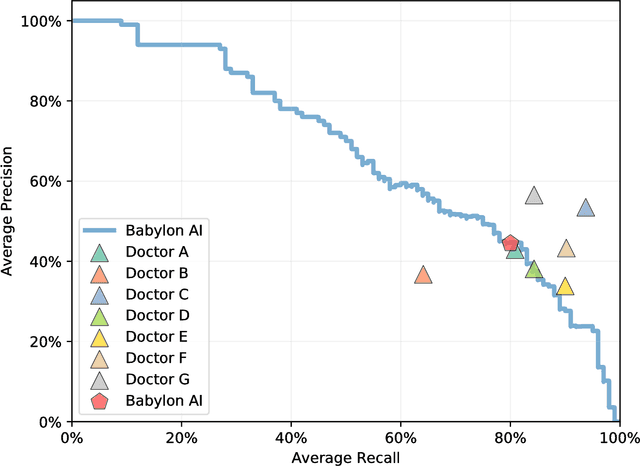
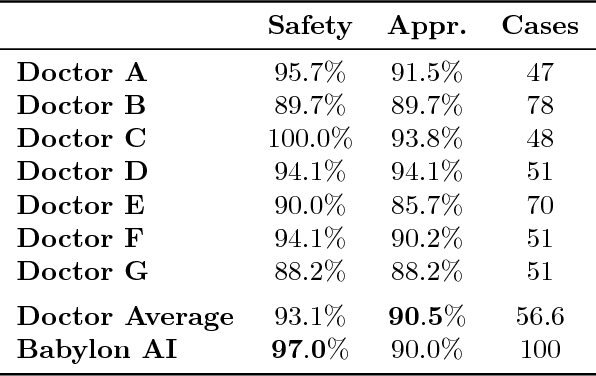
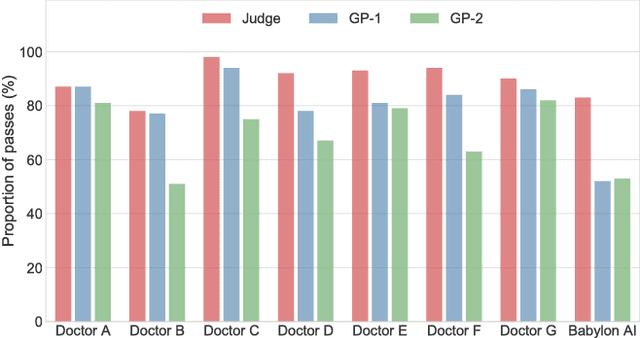
Abstract:Online symptom checkers have significant potential to improve patient care, however their reliability and accuracy remain variable. We hypothesised that an artificial intelligence (AI) powered triage and diagnostic system would compare favourably with human doctors with respect to triage and diagnostic accuracy. We performed a prospective validation study of the accuracy and safety of an AI powered triage and diagnostic system. Identical cases were evaluated by both an AI system and human doctors. Differential diagnoses and triage outcomes were evaluated by an independent judge, who was blinded from knowing the source (AI system or human doctor) of the outcomes. Independently of these cases, vignettes from publicly available resources were also assessed to provide a benchmark to previous studies and the diagnostic component of the MRCGP exam. Overall we found that the Babylon AI powered Triage and Diagnostic System was able to identify the condition modelled by a clinical vignette with accuracy comparable to human doctors (in terms of precision and recall). In addition, we found that the triage advice recommended by the AI System was, on average, safer than that of human doctors, when compared to the ranges of acceptable triage provided by independent expert judges, with only a minimal reduction in appropriateness.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge