Paul Elbers
Yet Another ICU Benchmark: A Flexible Multi-Center Framework for Clinical ML
Jun 08, 2023Abstract:Medical applications of machine learning (ML) have experienced a surge in popularity in recent years. The intensive care unit (ICU) is a natural habitat for ML given the abundance of available data from electronic health records. Models have been proposed to address numerous ICU prediction tasks like the early detection of complications. While authors frequently report state-of-the-art performance, it is challenging to verify claims of superiority. Datasets and code are not always published, and cohort definitions, preprocessing pipelines, and training setups are difficult to reproduce. This work introduces Yet Another ICU Benchmark (YAIB), a modular framework that allows researchers to define reproducible and comparable clinical ML experiments; we offer an end-to-end solution from cohort definition to model evaluation. The framework natively supports most open-access ICU datasets (MIMIC III/IV, eICU, HiRID, AUMCdb) and is easily adaptable to future ICU datasets. Combined with a transparent preprocessing pipeline and extensible training code for multiple ML and deep learning models, YAIB enables unified model development. Our benchmark comes with five predefined established prediction tasks (mortality, acute kidney injury, sepsis, kidney function, and length of stay) developed in collaboration with clinicians. Adding further tasks is straightforward by design. Using YAIB, we demonstrate that the choice of dataset, cohort definition, and preprocessing have a major impact on the prediction performance - often more so than model class - indicating an urgent need for YAIB as a holistic benchmarking tool. We provide our work to the clinical ML community to accelerate method development and enable real-world clinical implementations. Software Repository: https://github.com/rvandewater/YAIB.
Improving adaptability to new environments and removing catastrophic forgetting in Reinforcement Learning by using an eco-system of agents
Apr 13, 2022



Abstract:Adapting a Reinforcement Learning (RL) agent to an unseen environment is a difficult task due to typical over-fitting on the training environment. RL agents are often capable of solving environments very close to the trained environment, but when environments become substantially different, their performance quickly drops. When agents are retrained on new environments, a second issue arises: there is a risk of catastrophic forgetting, where the performance on previously seen environments is seriously hampered. This paper proposes a novel approach that exploits an ecosystem of agents to address both concerns. Hereby, the (limited) adaptive power of individual agents is harvested to build a highly adaptive ecosystem. This allows to transfer part of the workload from learning to inference. An evaluation of the approach on two distinct distributions of environments shows that our approach outperforms state-of-the-art techniques in terms of adaptability/generalization as well as avoids catastrophic forgetting.
Out-of-Distribution Detection for Medical Applications: Guidelines for Practical Evaluation
Sep 30, 2021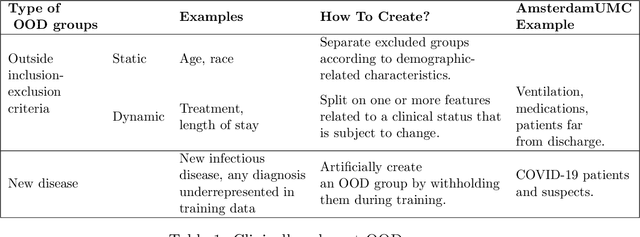
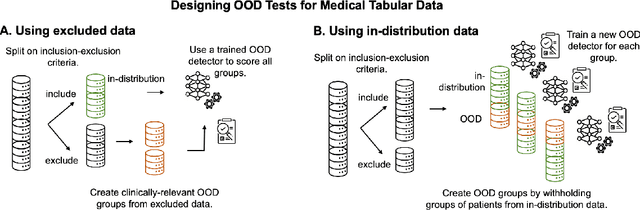
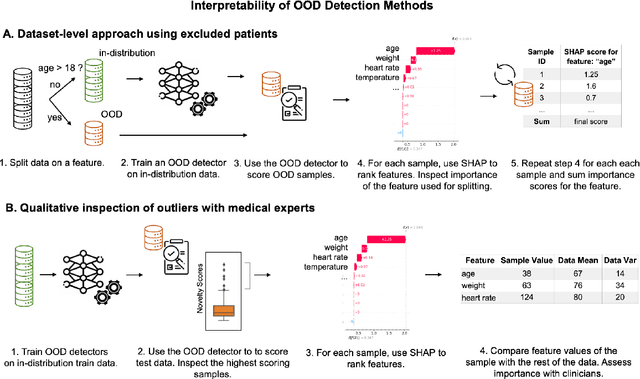
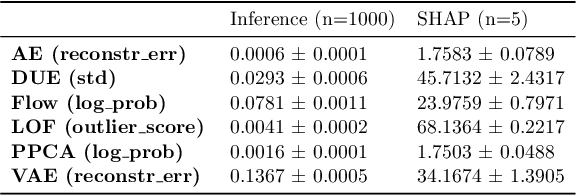
Abstract:Detection of Out-of-Distribution (OOD) samples in real time is a crucial safety check for deployment of machine learning models in the medical field. Despite a growing number of uncertainty quantification techniques, there is a lack of evaluation guidelines on how to select OOD detection methods in practice. This gap impedes implementation of OOD detection methods for real-world applications. Here, we propose a series of practical considerations and tests to choose the best OOD detector for a specific medical dataset. These guidelines are illustrated on a real-life use case of Electronic Health Records (EHR). Our results can serve as a guide for implementation of OOD detection methods in clinical practice, mitigating risks associated with the use of machine learning models in healthcare.
Integrating Expert ODEs into Neural ODEs: Pharmacology and Disease Progression
Jun 17, 2021



Abstract:Modeling a system's temporal behaviour in reaction to external stimuli is a fundamental problem in many areas. Pure Machine Learning (ML) approaches often fail in the small sample regime and cannot provide actionable insights beyond predictions. A promising modification has been to incorporate expert domain knowledge into ML models. The application we consider is predicting the progression of disease under medications, where a plethora of domain knowledge is available from pharmacology. Pharmacological models describe the dynamics of carefully-chosen medically meaningful variables in terms of systems of Ordinary Differential Equations (ODEs). However, these models only describe a limited collection of variables, and these variables are often not observable in clinical environments. To close this gap, we propose the latent hybridisation model (LHM) that integrates a system of expert-designed ODEs with machine-learned Neural ODEs to fully describe the dynamics of the system and to link the expert and latent variables to observable quantities. We evaluated LHM on synthetic data as well as real-world intensive care data of COVID-19 patients. LHM consistently outperforms previous works, especially when few training samples are available such as at the beginning of the pandemic.
Hide-and-Seek Privacy Challenge
Jul 24, 2020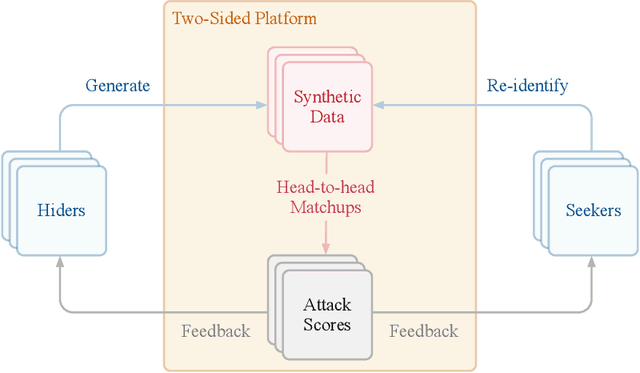
Abstract:The clinical time-series setting poses a unique combination of challenges to data modeling and sharing. Due to the high dimensionality of clinical time series, adequate de-identification to preserve privacy while retaining data utility is difficult to achieve using common de-identification techniques. An innovative approach to this problem is synthetic data generation. From a technical perspective, a good generative model for time-series data should preserve temporal dynamics, in the sense that new sequences respect the original relationships between high-dimensional variables across time. From the privacy perspective, the model should prevent patient re-identification by limiting vulnerability to membership inference attacks. The NeurIPS 2020 Hide-and-Seek Privacy Challenge is a novel two-tracked competition to simultaneously accelerate progress in tackling both problems. In our head-to-head format, participants in the synthetic data generation track (i.e. "hiders") and the patient re-identification track (i.e. "seekers") are directly pitted against each other by way of a new, high-quality intensive care time-series dataset: the AmsterdamUMCdb dataset. Ultimately, we seek to advance generative techniques for dense and high-dimensional temporal data streams that are (1) clinically meaningful in terms of fidelity and predictivity, as well as (2) capable of minimizing membership privacy risks in terms of the concrete notion of patient re-identification.
Bayesian Modelling in Practice: Using Uncertainty to Improve Trustworthiness in Medical Applications
Jun 20, 2019



Abstract:The Intensive Care Unit (ICU) is a hospital department where machine learning has the potential to provide valuable assistance in clinical decision making. Classical machine learning models usually only provide point-estimates and no uncertainty of predictions. In practice, uncertain predictions should be presented to doctors with extra care in order to prevent potentially catastrophic treatment decisions. In this work we show how Bayesian modelling and the predictive uncertainty that it provides can be used to mitigate risk of misguided prediction and to detect out-of-domain examples in a medical setting. We derive analytically a bound on the prediction loss with respect to predictive uncertainty. The bound shows that uncertainty can mitigate loss. Furthermore, we apply a Bayesian Neural Network to the MIMIC-III dataset, predicting risk of mortality of ICU patients. Our empirical results show that uncertainty can indeed prevent potential errors and reliably identifies out-of-domain patients. These results suggest that Bayesian predictive uncertainty can greatly improve trustworthiness of machine learning models in high-risk settings such as the ICU.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge