Danielle Belgrave
Consensus, dissensus and synergy between clinicians and specialist foundation models in radiology report generation
Dec 06, 2023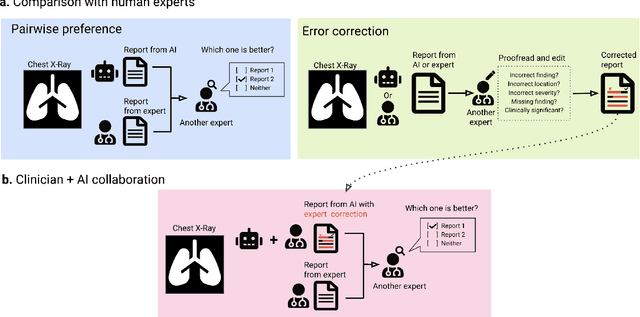
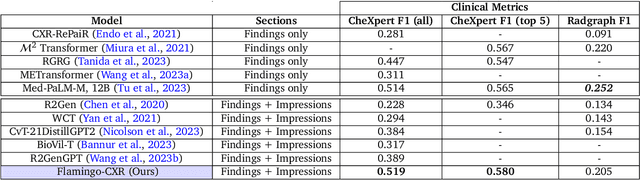
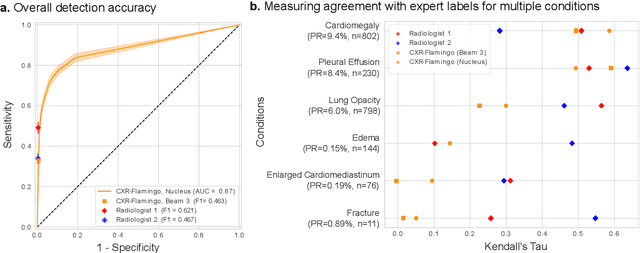
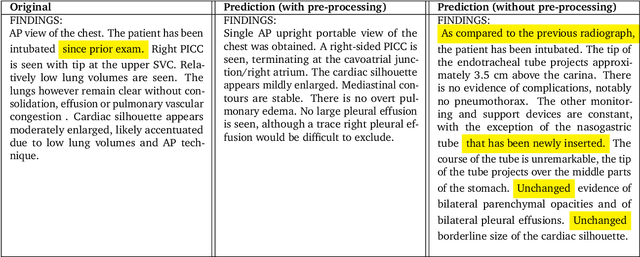
Abstract:Radiology reports are an instrumental part of modern medicine, informing key clinical decisions such as diagnosis and treatment. The worldwide shortage of radiologists, however, restricts access to expert care and imposes heavy workloads, contributing to avoidable errors and delays in report delivery. While recent progress in automated report generation with vision-language models offer clear potential in ameliorating the situation, the path to real-world adoption has been stymied by the challenge of evaluating the clinical quality of AI-generated reports. In this study, we build a state-of-the-art report generation system for chest radiographs, \textit{Flamingo-CXR}, by fine-tuning a well-known vision-language foundation model on radiology data. To evaluate the quality of the AI-generated reports, a group of 16 certified radiologists provide detailed evaluations of AI-generated and human written reports for chest X-rays from an intensive care setting in the United States and an inpatient setting in India. At least one radiologist (out of two per case) preferred the AI report to the ground truth report in over 60$\%$ of cases for both datasets. Amongst the subset of AI-generated reports that contain errors, the most frequently cited reasons were related to the location and finding, whereas for human written reports, most mistakes were related to severity and finding. This disparity suggested potential complementarity between our AI system and human experts, prompting us to develop an assistive scenario in which \textit{Flamingo-CXR} generates a first-draft report, which is subsequently revised by a clinician. This is the first demonstration of clinician-AI collaboration for report writing, and the resultant reports are assessed to be equivalent or preferred by at least one radiologist to reports written by experts alone in 80$\%$ of in-patient cases and 60$\%$ of intensive care cases.
Generative models improve fairness of medical classifiers under distribution shifts
Apr 18, 2023Abstract:A ubiquitous challenge in machine learning is the problem of domain generalisation. This can exacerbate bias against groups or labels that are underrepresented in the datasets used for model development. Model bias can lead to unintended harms, especially in safety-critical applications like healthcare. Furthermore, the challenge is compounded by the difficulty of obtaining labelled data due to high cost or lack of readily available domain expertise. In our work, we show that learning realistic augmentations automatically from data is possible in a label-efficient manner using generative models. In particular, we leverage the higher abundance of unlabelled data to capture the underlying data distribution of different conditions and subgroups for an imaging modality. By conditioning generative models on appropriate labels, we can steer the distribution of synthetic examples according to specific requirements. We demonstrate that these learned augmentations can surpass heuristic ones by making models more robust and statistically fair in- and out-of-distribution. To evaluate the generality of our approach, we study 3 distinct medical imaging contexts of varying difficulty: (i) histopathology images from a publicly available generalisation benchmark, (ii) chest X-rays from publicly available clinical datasets, and (iii) dermatology images characterised by complex shifts and imaging conditions. Complementing real training samples with synthetic ones improves the robustness of models in all three medical tasks and increases fairness by improving the accuracy of diagnosis within underrepresented groups. This approach leads to stark improvements OOD across modalities: 7.7% prediction accuracy improvement in histopathology, 5.2% in chest radiology with 44.6% lower fairness gap and a striking 63.5% improvement in high-risk sensitivity for dermatology with a 7.5x reduction in fairness gap.
Active Acquisition for Multimodal Temporal Data: A Challenging Decision-Making Task
Nov 09, 2022
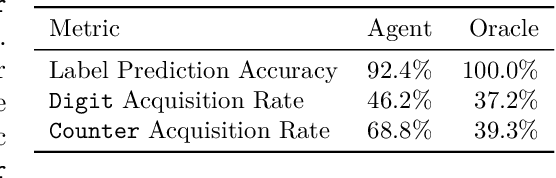
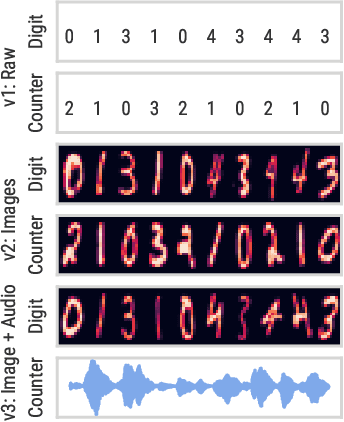

Abstract:We introduce a challenging decision-making task that we call active acquisition for multimodal temporal data (A2MT). In many real-world scenarios, input features are not readily available at test time and must instead be acquired at significant cost. With A2MT, we aim to learn agents that actively select which modalities of an input to acquire, trading off acquisition cost and predictive performance. A2MT extends a previous task called active feature acquisition to temporal decision making about high-dimensional inputs. Further, we propose a method based on the Perceiver IO architecture to address A2MT in practice. Our agents are able to solve a novel synthetic scenario requiring practically relevant cross-modal reasoning skills. On two large-scale, real-world datasets, Kinetics-700 and AudioSet, our agents successfully learn cost-reactive acquisition behavior. However, an ablation reveals they are unable to learn to learn adaptive acquisition strategies, emphasizing the difficulty of the task even for state-of-the-art models. Applications of A2MT may be impactful in domains like medicine, robotics, or finance, where modalities differ in acquisition cost and informativeness.
Causal Discovery for Causal Bandits utilizing Separating Sets
Sep 16, 2020



Abstract:The Causal Bandit is a variant of the classic Bandit problem where an agent must identify the best action in a sequential decision-making process, where the reward distribution of the actions displays a non-trivial dependence structure that is governed by a causal model. All methods proposed thus far in the literature rely on exact prior knowledge of the causal model to obtain improved estimators for the reward. We formulate a new causal bandit algorithm that is the first to no longer rely on explicit prior causal knowledge and instead uses the output of causal discovery algorithms. This algorithm relies on a new estimator based on separating sets, a causal structure already known in causal discovery literature. We show that given a separating set, this estimator is unbiased, and has lower variance compared to the sample mean. We derive a concentration bound and construct a UCB-type algorithm based on this bound, as well as a Thompson sampling variant. We compare our algorithms with traditional bandit algorithms on simulation data. On these problems, our algorithms show a significant boost in performance.
Hide-and-Seek Privacy Challenge
Jul 24, 2020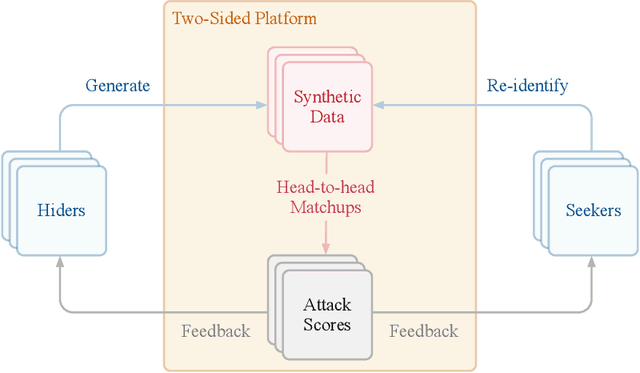
Abstract:The clinical time-series setting poses a unique combination of challenges to data modeling and sharing. Due to the high dimensionality of clinical time series, adequate de-identification to preserve privacy while retaining data utility is difficult to achieve using common de-identification techniques. An innovative approach to this problem is synthetic data generation. From a technical perspective, a good generative model for time-series data should preserve temporal dynamics, in the sense that new sequences respect the original relationships between high-dimensional variables across time. From the privacy perspective, the model should prevent patient re-identification by limiting vulnerability to membership inference attacks. The NeurIPS 2020 Hide-and-Seek Privacy Challenge is a novel two-tracked competition to simultaneously accelerate progress in tackling both problems. In our head-to-head format, participants in the synthetic data generation track (i.e. "hiders") and the patient re-identification track (i.e. "seekers") are directly pitted against each other by way of a new, high-quality intensive care time-series dataset: the AmsterdamUMCdb dataset. Ultimately, we seek to advance generative techniques for dense and high-dimensional temporal data streams that are (1) clinically meaningful in terms of fidelity and predictivity, as well as (2) capable of minimizing membership privacy risks in terms of the concrete notion of patient re-identification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge