Abhijit Guha Roy
MINT: A wrapper to make multi-modal and multi-image AI models interactive
Jan 22, 2024Abstract:During the diagnostic process, doctors incorporate multimodal information including imaging and the medical history - and similarly medical AI development has increasingly become multimodal. In this paper we tackle a more subtle challenge: doctors take a targeted medical history to obtain only the most pertinent pieces of information; how do we enable AI to do the same? We develop a wrapper method named MINT (Make your model INTeractive) that automatically determines what pieces of information are most valuable at each step, and ask for only the most useful information. We demonstrate the efficacy of MINT wrapping a skin disease prediction model, where multiple images and a set of optional answers to $25$ standard metadata questions (i.e., structured medical history) are used by a multi-modal deep network to provide a differential diagnosis. We show that MINT can identify whether metadata inputs are needed and if so, which question to ask next. We also demonstrate that when collecting multiple images, MINT can identify if an additional image would be beneficial, and if so, which type of image to capture. We showed that MINT reduces the number of metadata and image inputs needed by 82% and 36.2% respectively, while maintaining predictive performance. Using real-world AI dermatology system data, we show that needing fewer inputs can retain users that may otherwise fail to complete the system submission and drop off without a diagnosis. Qualitative examples show MINT can closely mimic the step-by-step decision making process of a clinical workflow and how this is different for straight forward cases versus more difficult, ambiguous cases. Finally we demonstrate how MINT is robust to different underlying multi-model classifiers and can be easily adapted to user requirements without significant model re-training.
Conformal prediction under ambiguous ground truth
Jul 18, 2023



Abstract:In safety-critical classification tasks, conformal prediction allows to perform rigorous uncertainty quantification by providing confidence sets including the true class with a user-specified probability. This generally assumes the availability of a held-out calibration set with access to ground truth labels. Unfortunately, in many domains, such labels are difficult to obtain and usually approximated by aggregating expert opinions. In fact, this holds true for almost all datasets, including well-known ones such as CIFAR and ImageNet. Applying conformal prediction using such labels underestimates uncertainty. Indeed, when expert opinions are not resolvable, there is inherent ambiguity present in the labels. That is, we do not have ``crisp'', definitive ground truth labels and this uncertainty should be taken into account during calibration. In this paper, we develop a conformal prediction framework for such ambiguous ground truth settings which relies on an approximation of the underlying posterior distribution of labels given inputs. We demonstrate our methodology on synthetic and real datasets, including a case study of skin condition classification in dermatology.
Evaluating AI systems under uncertain ground truth: a case study in dermatology
Jul 05, 2023



Abstract:For safety, AI systems in health undergo thorough evaluations before deployment, validating their predictions against a ground truth that is assumed certain. However, this is actually not the case and the ground truth may be uncertain. Unfortunately, this is largely ignored in standard evaluation of AI models but can have severe consequences such as overestimating the future performance. To avoid this, we measure the effects of ground truth uncertainty, which we assume decomposes into two main components: annotation uncertainty which stems from the lack of reliable annotations, and inherent uncertainty due to limited observational information. This ground truth uncertainty is ignored when estimating the ground truth by deterministically aggregating annotations, e.g., by majority voting or averaging. In contrast, we propose a framework where aggregation is done using a statistical model. Specifically, we frame aggregation of annotations as posterior inference of so-called plausibilities, representing distributions over classes in a classification setting, subject to a hyper-parameter encoding annotator reliability. Based on this model, we propose a metric for measuring annotation uncertainty and provide uncertainty-adjusted metrics for performance evaluation. We present a case study applying our framework to skin condition classification from images where annotations are provided in the form of differential diagnoses. The deterministic adjudication process called inverse rank normalization (IRN) from previous work ignores ground truth uncertainty in evaluation. Instead, we present two alternative statistical models: a probabilistic version of IRN and a Plackett-Luce-based model. We find that a large portion of the dataset exhibits significant ground truth uncertainty and standard IRN-based evaluation severely over-estimates performance without providing uncertainty estimates.
Generative models improve fairness of medical classifiers under distribution shifts
Apr 18, 2023Abstract:A ubiquitous challenge in machine learning is the problem of domain generalisation. This can exacerbate bias against groups or labels that are underrepresented in the datasets used for model development. Model bias can lead to unintended harms, especially in safety-critical applications like healthcare. Furthermore, the challenge is compounded by the difficulty of obtaining labelled data due to high cost or lack of readily available domain expertise. In our work, we show that learning realistic augmentations automatically from data is possible in a label-efficient manner using generative models. In particular, we leverage the higher abundance of unlabelled data to capture the underlying data distribution of different conditions and subgroups for an imaging modality. By conditioning generative models on appropriate labels, we can steer the distribution of synthetic examples according to specific requirements. We demonstrate that these learned augmentations can surpass heuristic ones by making models more robust and statistically fair in- and out-of-distribution. To evaluate the generality of our approach, we study 3 distinct medical imaging contexts of varying difficulty: (i) histopathology images from a publicly available generalisation benchmark, (ii) chest X-rays from publicly available clinical datasets, and (iii) dermatology images characterised by complex shifts and imaging conditions. Complementing real training samples with synthetic ones improves the robustness of models in all three medical tasks and increases fairness by improving the accuracy of diagnosis within underrepresented groups. This approach leads to stark improvements OOD across modalities: 7.7% prediction accuracy improvement in histopathology, 5.2% in chest radiology with 44.6% lower fairness gap and a striking 63.5% improvement in high-risk sensitivity for dermatology with a 7.5x reduction in fairness gap.
Robust and Efficient Medical Imaging with Self-Supervision
May 19, 2022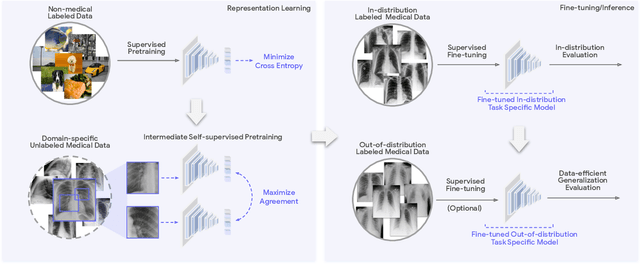
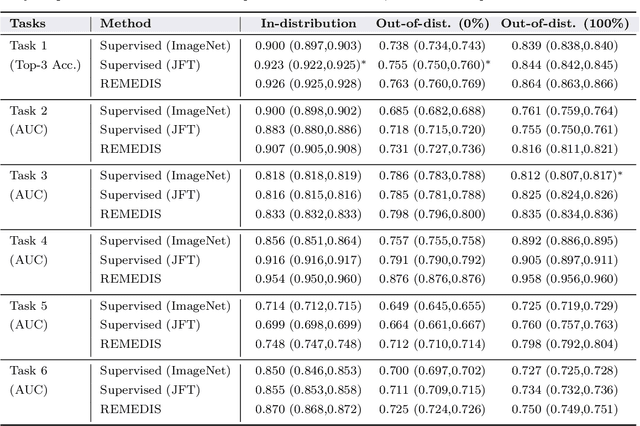
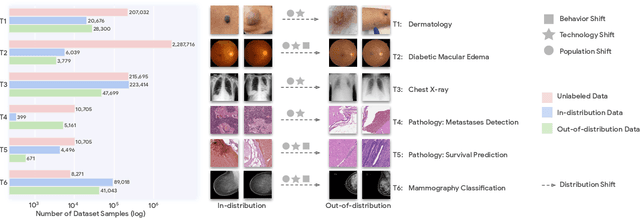
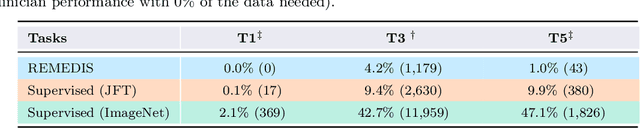
Abstract:Recent progress in Medical Artificial Intelligence (AI) has delivered systems that can reach clinical expert level performance. However, such systems tend to demonstrate sub-optimal "out-of-distribution" performance when evaluated in clinical settings different from the training environment. A common mitigation strategy is to develop separate systems for each clinical setting using site-specific data [1]. However, this quickly becomes impractical as medical data is time-consuming to acquire and expensive to annotate [2]. Thus, the problem of "data-efficient generalization" presents an ongoing difficulty for Medical AI development. Although progress in representation learning shows promise, their benefits have not been rigorously studied, specifically for out-of-distribution settings. To meet these challenges, we present REMEDIS, a unified representation learning strategy to improve robustness and data-efficiency of medical imaging AI. REMEDIS uses a generic combination of large-scale supervised transfer learning with self-supervised learning and requires little task-specific customization. We study a diverse range of medical imaging tasks and simulate three realistic application scenarios using retrospective data. REMEDIS exhibits significantly improved in-distribution performance with up to 11.5% relative improvement in diagnostic accuracy over a strong supervised baseline. More importantly, our strategy leads to strong data-efficient generalization of medical imaging AI, matching strong supervised baselines using between 1% to 33% of retraining data across tasks. These results suggest that REMEDIS can significantly accelerate the life-cycle of medical imaging AI development thereby presenting an important step forward for medical imaging AI to deliver broad impact.
A Simple Fix to Mahalanobis Distance for Improving Near-OOD Detection
Jun 16, 2021



Abstract:Mahalanobis distance (MD) is a simple and popular post-processing method for detecting out-of-distribution (OOD) inputs in neural networks. We analyze its failure modes for near-OOD detection and propose a simple fix called relative Mahalanobis distance (RMD) which improves performance and is more robust to hyperparameter choice. On a wide selection of challenging vision, language, and biology OOD benchmarks (CIFAR-100 vs CIFAR-10, CLINC OOD intent detection, Genomics OOD), we show that RMD meaningfully improves upon MD performance (by up to 15% AUROC on genomics OOD).
Does Your Dermatology Classifier Know What It Doesn't Know? Detecting the Long-Tail of Unseen Conditions
Apr 08, 2021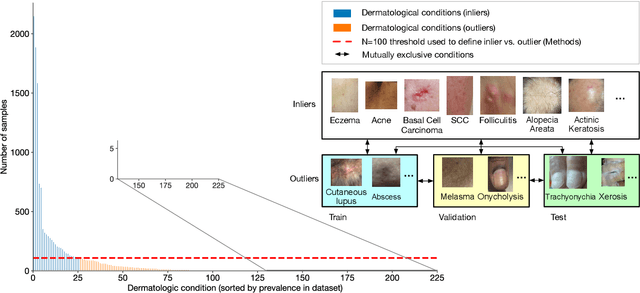
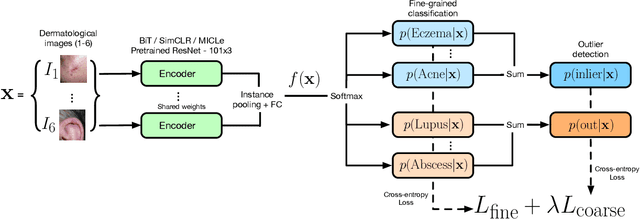
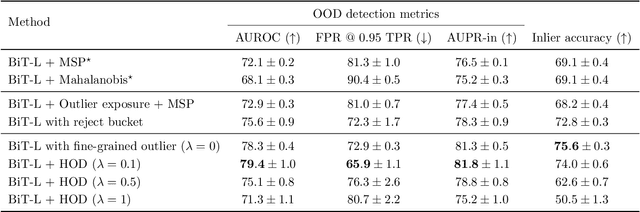
Abstract:We develop and rigorously evaluate a deep learning based system that can accurately classify skin conditions while detecting rare conditions for which there is not enough data available for training a confident classifier. We frame this task as an out-of-distribution (OOD) detection problem. Our novel approach, hierarchical outlier detection (HOD) assigns multiple abstention classes for each training outlier class and jointly performs a coarse classification of inliers vs. outliers, along with fine-grained classification of the individual classes. We demonstrate the effectiveness of the HOD loss in conjunction with modern representation learning approaches (BiT, SimCLR, MICLe) and explore different ensembling strategies for further improving the results. We perform an extensive subgroup analysis over conditions of varying risk levels and different skin types to investigate how the OOD detection performance changes over each subgroup and demonstrate the gains of our framework in comparison to baselines. Finally, we introduce a cost metric to approximate downstream clinical impact. We use this cost metric to compare the proposed method against a baseline system, thereby making a stronger case for the overall system effectiveness in a real-world deployment scenario.
Contrastive Training for Improved Out-of-Distribution Detection
Jul 10, 2020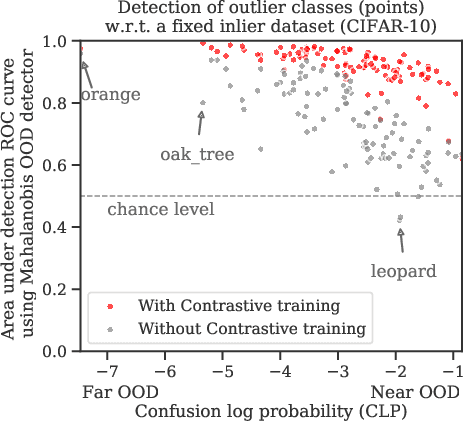
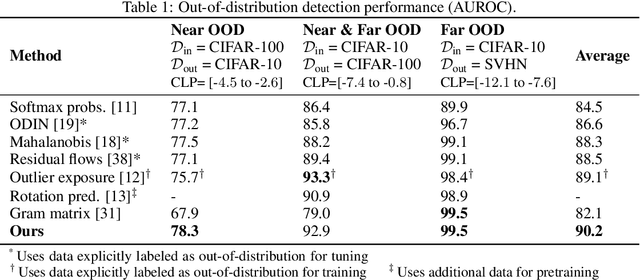
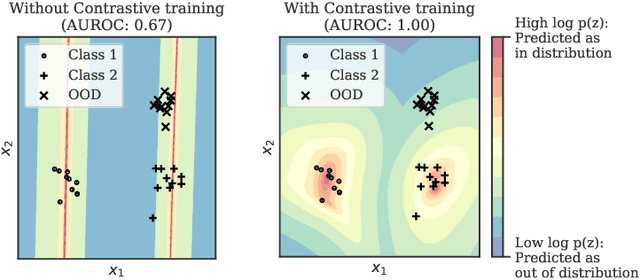
Abstract:Reliable detection of out-of-distribution (OOD) inputs is increasingly understood to be a precondition for deployment of machine learning systems. This paper proposes and investigates the use of contrastive training to boost OOD detection performance. Unlike leading methods for OOD detection, our approach does not require access to examples labeled explicitly as OOD, which can be difficult to collect in practice. We show in extensive experiments that contrastive training significantly helps OOD detection performance on a number of common benchmarks. By introducing and employing the Confusion Log Probability (CLP) score, which quantifies the difficulty of the OOD detection task by capturing the similarity of inlier and outlier datasets, we show that our method especially improves performance in the `near OOD' classes -- a particularly challenging setting for previous methods.
Importance Driven Continual Learning for Segmentation Across Domains
Apr 30, 2020
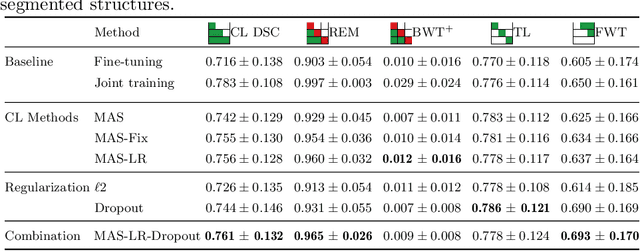


Abstract:The ability of neural networks to continuously learn and adapt to new tasks while retaining prior knowledge is crucial for many applications. However, current neural networks tend to forget previously learned tasks when trained on new ones, i.e., they suffer from Catastrophic Forgetting (CF). The objective of Continual Learning (CL) is to alleviate this problem, which is particularly relevant for medical applications, where it may not be feasible to store and access previously used sensitive patient data. In this work, we propose a Continual Learning approach for brain segmentation, where a single network is consecutively trained on samples from different domains. We build upon an importance driven approach and adapt it for medical image segmentation. Particularly, we introduce learning rate regularization to prevent the loss of the network's knowledge. Our results demonstrate that directly restricting the adaptation of important network parameters clearly reduces Catastrophic Forgetting for segmentation across domains.
Recalibrating 3D ConvNets with Project & Excite
Feb 25, 2020



Abstract:Fully Convolutional Neural Networks (F-CNNs) achieve state-of-the-art performance for segmentation tasks in computer vision and medical imaging. Recently, computational blocks termed squeeze and excitation (SE) have been introduced to recalibrate F-CNN feature maps both channel- and spatial-wise, boosting segmentation performance while only minimally increasing the model complexity. So far, the development of SE blocks has focused on 2D architectures. For volumetric medical images, however, 3D F-CNNs are a natural choice. In this article, we extend existing 2D recalibration methods to 3D and propose a generic compress-process-recalibrate pipeline for easy comparison of such blocks. We further introduce Project & Excite (PE) modules, customized for 3D networks. In contrast to existing modules, Project \& Excite does not perform global average pooling but compresses feature maps along different spatial dimensions of the tensor separately to retain more spatial information that is subsequently used in the excitation step. We evaluate the modules on two challenging tasks, whole-brain segmentation of MRI scans and whole-body segmentation of CT scans. We demonstrate that PE modules can be easily integrated into 3D F-CNNs, boosting performance up to 0.3 in Dice Score and outperforming 3D extensions of other recalibration blocks, while only marginally increasing the model complexity. Our code is publicly available on https://github.com/ai-med/squeeze_and_excitation .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge