Anne-Marie Rickmann
Using Foundation Models as Pseudo-Label Generators for Pre-Clinical 4D Cardiac CT Segmentation
May 14, 2025Abstract:Cardiac image segmentation is an important step in many cardiac image analysis and modeling tasks such as motion tracking or simulations of cardiac mechanics. While deep learning has greatly advanced segmentation in clinical settings, there is limited work on pre-clinical imaging, notably in porcine models, which are often used due to their anatomical and physiological similarity to humans. However, differences between species create a domain shift that complicates direct model transfer from human to pig data. Recently, foundation models trained on large human datasets have shown promise for robust medical image segmentation; yet their applicability to porcine data remains largely unexplored. In this work, we investigate whether foundation models can generate sufficiently accurate pseudo-labels for pig cardiac CT and propose a simple self-training approach to iteratively refine these labels. Our method requires no manually annotated pig data, relying instead on iterative updates to improve segmentation quality. We demonstrate that this self-training process not only enhances segmentation accuracy but also smooths out temporal inconsistencies across consecutive frames. Although our results are encouraging, there remains room for improvement, for example by incorporating more sophisticated self-training strategies and by exploring additional foundation models and other cardiac imaging technologies.
Progressive Test Time Energy Adaptation for Medical Image Segmentation
Mar 20, 2025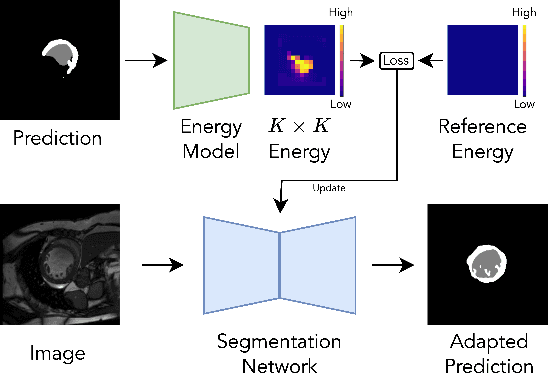
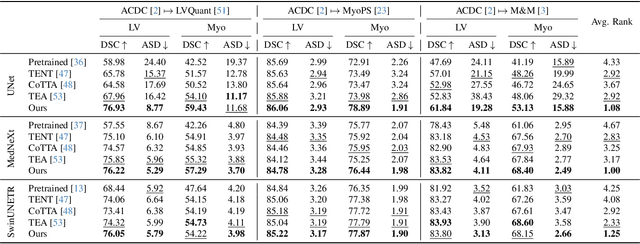
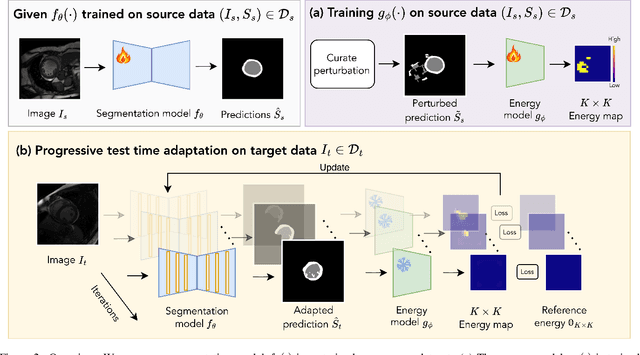
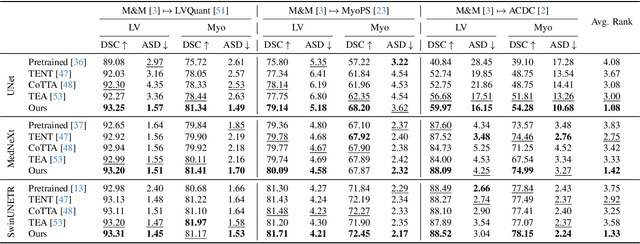
Abstract:We propose a model-agnostic, progressive test-time energy adaptation approach for medical image segmentation. Maintaining model performance across diverse medical datasets is challenging, as distribution shifts arise from inconsistent imaging protocols and patient variations. Unlike domain adaptation methods that require multiple passes through target data - impractical in clinical settings - our approach adapts pretrained models progressively as they process test data. Our method leverages a shape energy model trained on source data, which assigns an energy score at the patch level to segmentation maps: low energy represents in-distribution (accurate) shapes, while high energy signals out-of-distribution (erroneous) predictions. By minimizing this energy score at test time, we refine the segmentation model to align with the target distribution. To validate the effectiveness and adaptability, we evaluated our framework on eight public MRI (bSSFP, T1- and T2-weighted) and X-ray datasets spanning cardiac, spinal cord, and lung segmentation. We consistently outperform baselines both quantitatively and qualitatively.
V2C-Long: Longitudinal Cortex Reconstruction with Spatiotemporal Correspondence
Feb 27, 2024



Abstract:Reconstructing the cortex from longitudinal MRI is indispensable for analyzing morphological changes in the human brain. Despite the recent disruption of cortical surface reconstruction with deep learning, challenges arising from longitudinal data are still persistent. Especially the lack of strong spatiotemporal point correspondence hinders downstream analyses due to the introduced noise. To address this issue, we present V2C-Long, the first dedicated deep learning-based cortex reconstruction method for longitudinal MRI. In contrast to existing methods, V2C-Long surfaces are directly comparable in a cross-sectional and longitudinal manner. We establish strong inherent spatiotemporal correspondences via a novel composition of two deep mesh deformation networks and fast aggregation of feature-enhanced within-subject templates. The results on internal and external test data demonstrate that V2C-Long yields cortical surfaces with improved accuracy and consistency compared to previous methods. Finally, this improvement manifests in higher sensitivity to regional cortical atrophy in Alzheimer's disease.
Neural deformation fields for template-based reconstruction of cortical surfaces from MRI
Jan 23, 2024



Abstract:The reconstruction of cortical surfaces is a prerequisite for quantitative analyses of the cerebral cortex in magnetic resonance imaging (MRI). Existing segmentation-based methods separate the surface registration from the surface extraction, which is computationally inefficient and prone to distortions. We introduce Vox2Cortex-Flow (V2C-Flow), a deep mesh-deformation technique that learns a deformation field from a brain template to the cortical surfaces of an MRI scan. To this end, we present a geometric neural network that models the deformation-describing ordinary differential equation in a continuous manner. The network architecture comprises convolutional and graph-convolutional layers, which allows it to work with images and meshes at the same time. V2C-Flow is not only very fast, requiring less than two seconds to infer all four cortical surfaces, but also establishes vertex-wise correspondences to the template during reconstruction. In addition, V2C-Flow is the first approach for cortex reconstruction that models white matter and pial surfaces jointly, therefore avoiding intersections between them. Our comprehensive experiments on internal and external test data demonstrate that V2C-Flow results in cortical surfaces that are state-of-the-art in terms of accuracy. Moreover, we show that the established correspondences are more consistent than in FreeSurfer and that they can directly be utilized for cortex parcellation and group analyses of cortical thickness.
Keep the Faith: Faithful Explanations in Convolutional Neural Networks for Case-Based Reasoning
Dec 19, 2023


Abstract:Explaining predictions of black-box neural networks is crucial when applied to decision-critical tasks. Thus, attribution maps are commonly used to identify important image regions, despite prior work showing that humans prefer explanations based on similar examples. To this end, ProtoPNet learns a set of class-representative feature vectors (prototypes) for case-based reasoning. During inference, similarities of latent features to prototypes are linearly classified to form predictions and attribution maps are provided to explain the similarity. In this work, we evaluate whether architectures for case-based reasoning fulfill established axioms required for faithful explanations using the example of ProtoPNet. We show that such architectures allow the extraction of faithful explanations. However, we prove that the attribution maps used to explain the similarities violate the axioms. We propose a new procedure to extract explanations for trained ProtoPNets, named ProtoPFaith. Conceptually, these explanations are Shapley values, calculated on the similarity scores of each prototype. They allow to faithfully answer which prototypes are present in an unseen image and quantify each pixel's contribution to that presence, thereby complying with all axioms. The theoretical violations of ProtoPNet manifest in our experiments on three datasets (CUB-200-2011, Stanford Dogs, RSNA) and five architectures (ConvNet, ResNet, ResNet50, WideResNet50, ResNeXt50). Our experiments show a qualitative difference between the explanations given by ProtoPNet and ProtoPFaith. Additionally, we quantify the explanations with the Area Over the Perturbation Curve, on which ProtoPFaith outperforms ProtoPNet on all experiments by a factor $>10^3$.
Meshes Meet Voxels: Abdominal Organ Segmentation via Diffeomorphic Deformations
Jun 27, 2023Abstract:Abdominal multi-organ segmentation from CT and MRI is an essential prerequisite for surgical planning and computer-aided navigation systems. Three-dimensional numeric representations of abdominal shapes are further important for quantitative and statistical analyses thereof. Existing methods in the field, however, are unable to extract highly accurate 3D representations that are smooth, topologically correct, and match points on a template. In this work, we present UNetFlow, a novel diffeomorphic shape deformation approach for abdominal organs. UNetFlow combines the advantages of voxel-based and mesh-based approaches for 3D shape extraction. Our results demonstrate high accuracy with respect to manually annotated CT data and better topological correctness compared to previous methods. In addition, we show the generalization of UNetFlow to MRI.
HALOS: Hallucination-free Organ Segmentation after Organ Resection Surgery
Mar 14, 2023Abstract:The wide range of research in deep learning-based medical image segmentation pushed the boundaries in a multitude of applications. A clinically relevant problem that received less attention is the handling of scans with irregular anatomy, e.g., after organ resection. State-of-the-art segmentation models often lead to organ hallucinations, i.e., false-positive predictions of organs, which cannot be alleviated by oversampling or post-processing. Motivated by the increasing need to develop robust deep learning models, we propose HALOS for abdominal organ segmentation in MR images that handles cases after organ resection surgery. To this end, we combine missing organ classification and multi-organ segmentation tasks into a multi-task model, yielding a classification-assisted segmentation pipeline. The segmentation network learns to incorporate knowledge about organ existence via feature fusion modules. Extensive experiments on a small labeled test set and large-scale UK Biobank data demonstrate the effectiveness of our approach in terms of higher segmentation Dice scores and near-to-zero false positive prediction rate.
Joint Reconstruction and Parcellation of Cortical Surfaces
Sep 19, 2022
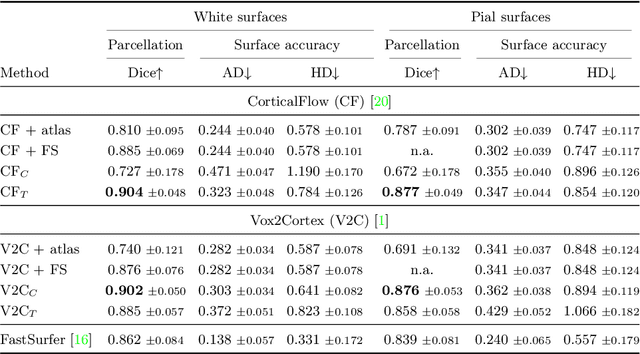

Abstract:The reconstruction of cerebral cortex surfaces from brain MRI scans is instrumental for the analysis of brain morphology and the detection of cortical thinning in neurodegenerative diseases like Alzheimer's disease (AD). Moreover, for a fine-grained analysis of atrophy patterns, the parcellation of the cortical surfaces into individual brain regions is required. For the former task, powerful deep learning approaches, which provide highly accurate brain surfaces of tissue boundaries from input MRI scans in seconds, have recently been proposed. However, these methods do not come with the ability to provide a parcellation of the reconstructed surfaces. Instead, separate brain-parcellation methods have been developed, which typically consider the cortical surfaces as given, often computed beforehand with FreeSurfer. In this work, we propose two options, one based on a graph classification branch and another based on a novel generic 3D reconstruction loss, to augment template-deformation algorithms such that the surface meshes directly come with an atlas-based brain parcellation. By combining both options with two of the latest cortical surface reconstruction algorithms, we attain highly accurate parcellations with a Dice score of 90.2 (graph classification branch) and 90.4 (novel reconstruction loss) together with state-of-the-art surfaces.
Vox2Cortex: Fast Explicit Reconstruction of Cortical Surfaces from 3D MRI Scans with Geometric Deep Neural Networks
Mar 18, 2022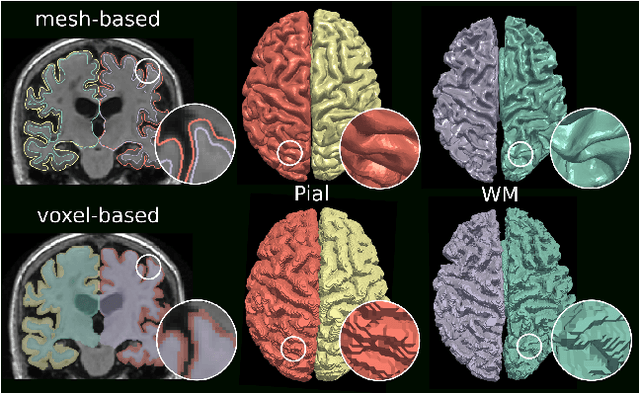
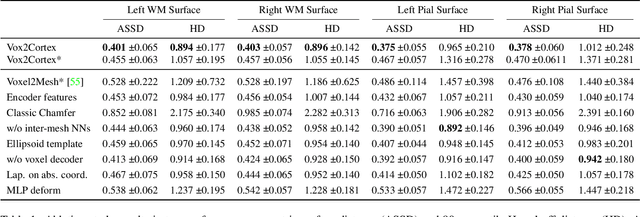

Abstract:The reconstruction of cortical surfaces from brain magnetic resonance imaging (MRI) scans is essential for quantitative analyses of cortical thickness and sulcal morphology. Although traditional and deep learning-based algorithmic pipelines exist for this purpose, they have two major drawbacks: lengthy runtimes of multiple hours (traditional) or intricate post-processing, such as mesh extraction and topology correction (deep learning-based). In this work, we address both of these issues and propose Vox2Cortex, a deep learning-based algorithm that directly yields topologically correct, three-dimensional meshes of the boundaries of the cortex. Vox2Cortex leverages convolutional and graph convolutional neural networks to deform an initial template to the densely folded geometry of the cortex represented by an input MRI scan. We show in extensive experiments on three brain MRI datasets that our meshes are as accurate as the ones reconstructed by state-of-the-art methods in the field, without the need for time- and resource-intensive post-processing. To accurately reconstruct the tightly folded cortex, we work with meshes containing about 168,000 vertices at test time, scaling deep explicit reconstruction methods to a new level.
STRUDEL: Self-Training with Uncertainty Dependent Label Refinement across Domains
Apr 23, 2021



Abstract:We propose an unsupervised domain adaptation (UDA) approach for white matter hyperintensity (WMH) segmentation, which uses Self-Training with Uncertainty DEpendent Label refinement (STRUDEL). Self-training has recently been introduced as a highly effective method for UDA, which is based on self-generated pseudo labels. However, pseudo labels can be very noisy and therefore deteriorate model performance. We propose to predict the uncertainty of pseudo labels and integrate it in the training process with an uncertainty-guided loss function to highlight labels with high certainty. STRUDEL is further improved by incorporating the segmentation output of an existing method in the pseudo label generation that showed high robustness for WMH segmentation. In our experiments, we evaluate STRUDEL with a standard U-Net and a modified network with a higher receptive field. Our results on WMH segmentation across datasets demonstrate the significant improvement of STRUDEL with respect to standard self-training.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge