Ira Ktena
Dj
Benchmarking Diversity in Image Generation via Attribute-Conditional Human Evaluation
Nov 13, 2025



Abstract:Despite advances in generation quality, current text-to-image (T2I) models often lack diversity, generating homogeneous outputs. This work introduces a framework to address the need for robust diversity evaluation in T2I models. Our framework systematically assesses diversity by evaluating individual concepts and their relevant factors of variation. Key contributions include: (1) a novel human evaluation template for nuanced diversity assessment; (2) a curated prompt set covering diverse concepts with their identified factors of variation (e.g. prompt: An image of an apple, factor of variation: color); and (3) a methodology for comparing models in terms of human annotations via binomial tests. Furthermore, we rigorously compare various image embeddings for diversity measurement. Notably, our principled approach enables ranking of T2I models by diversity, identifying categories where they particularly struggle. This research offers a robust methodology and insights, paving the way for improvements in T2I model diversity and metric development.
Dynamic Classifier-Free Diffusion Guidance via Online Feedback
Sep 19, 2025



Abstract:Classifier-free guidance (CFG) is a cornerstone of text-to-image diffusion models, yet its effectiveness is limited by the use of static guidance scales. This "one-size-fits-all" approach fails to adapt to the diverse requirements of different prompts; moreover, prior solutions like gradient-based correction or fixed heuristic schedules introduce additional complexities and fail to generalize. In this work, we challeng this static paradigm by introducing a framework for dynamic CFG scheduling. Our method leverages online feedback from a suite of general-purpose and specialized small-scale latent-space evaluations, such as CLIP for alignment, a discriminator for fidelity and a human preference reward model, to assess generation quality at each step of the reverse diffusion process. Based on this feedback, we perform a greedy search to select the optimal CFG scale for each timestep, creating a unique guidance schedule tailored to every prompt and sample. We demonstrate the effectiveness of our approach on both small-scale models and the state-of-the-art Imagen 3, showing significant improvements in text alignment, visual quality, text rendering and numerical reasoning. Notably, when compared against the default Imagen 3 baseline, our method achieves up to 53.8% human preference win-rate for overall preference, a figure that increases up to to 55.5% on prompts targeting specific capabilities like text rendering. Our work establishes that the optimal guidance schedule is inherently dynamic and prompt-dependent, and provides an efficient and generalizable framework to achieve it.
On the Risk of Misleading Reports: Diagnosing Textual Biases in Multimodal Clinical AI
Jul 31, 2025



Abstract:Clinical decision-making relies on the integrated analysis of medical images and the associated clinical reports. While Vision-Language Models (VLMs) can offer a unified framework for such tasks, they can exhibit strong biases toward one modality, frequently overlooking critical visual cues in favor of textual information. In this work, we introduce Selective Modality Shifting (SMS), a perturbation-based approach to quantify a model's reliance on each modality in binary classification tasks. By systematically swapping images or text between samples with opposing labels, we expose modality-specific biases. We assess six open-source VLMs-four generalist models and two fine-tuned for medical data-on two medical imaging datasets with distinct modalities: MIMIC-CXR (chest X-ray) and FairVLMed (scanning laser ophthalmoscopy). By assessing model performance and the calibration of every model in both unperturbed and perturbed settings, we reveal a marked dependency on text input, which persists despite the presence of complementary visual information. We also perform a qualitative attention-based analysis which further confirms that image content is often overshadowed by text details. Our findings highlight the importance of designing and evaluating multimodal medical models that genuinely integrate visual and textual cues, rather than relying on single-modality signals.
Imagen 3
Aug 13, 2024Abstract:We introduce Imagen 3, a latent diffusion model that generates high quality images from text prompts. We describe our quality and responsibility evaluations. Imagen 3 is preferred over other state-of-the-art (SOTA) models at the time of evaluation. In addition, we discuss issues around safety and representation, as well as methods we used to minimize the potential harm of our models.
FunBO: Discovering Acquisition Functions for Bayesian Optimization with FunSearch
Jun 07, 2024



Abstract:The sample efficiency of Bayesian optimization algorithms depends on carefully crafted acquisition functions (AFs) guiding the sequential collection of function evaluations. The best-performing AF can vary significantly across optimization problems, often requiring ad-hoc and problem-specific choices. This work tackles the challenge of designing novel AFs that perform well across a variety of experimental settings. Based on FunSearch, a recent work using Large Language Models (LLMs) for discovery in mathematical sciences, we propose FunBO, an LLM-based method that can be used to learn new AFs written in computer code by leveraging access to a limited number of evaluations for a set of objective functions. We provide the analytic expression of all discovered AFs and evaluate them on various global optimization benchmarks and hyperparameter optimization tasks. We show how FunBO identifies AFs that generalize well in and out of the training distribution of functions, thus outperforming established general-purpose AFs and achieving competitive performance against AFs that are customized to specific function types and are learned via transfer-learning algorithms.
Advancing Multimodal Medical Capabilities of Gemini
May 06, 2024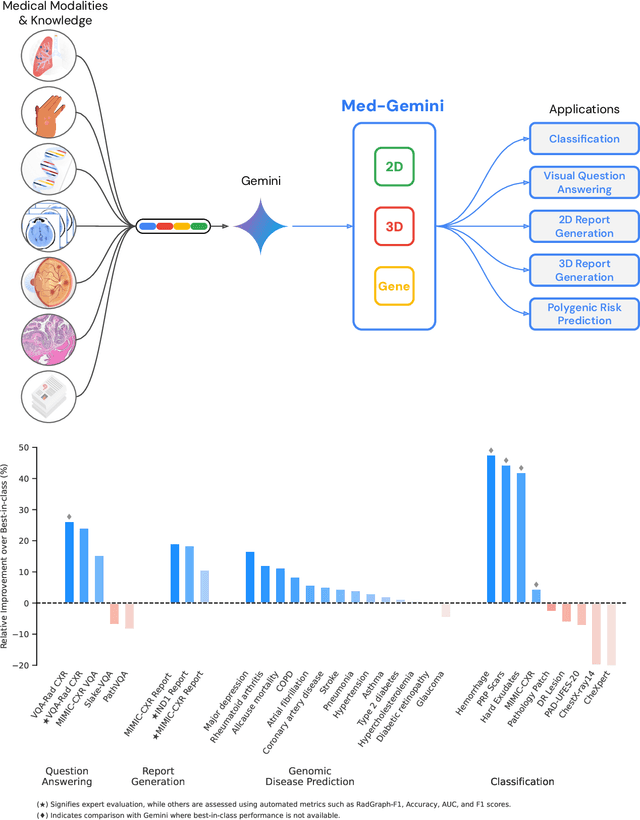
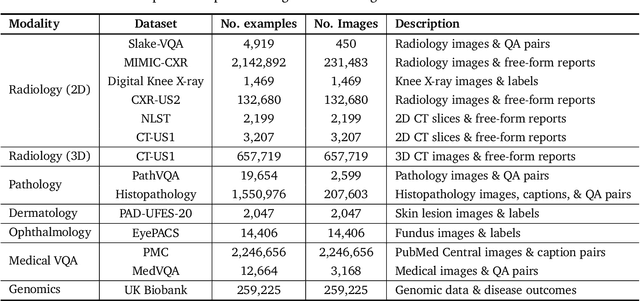

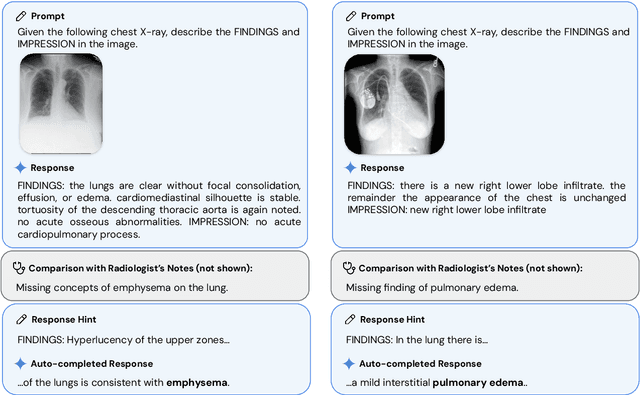
Abstract:Many clinical tasks require an understanding of specialized data, such as medical images and genomics, which is not typically found in general-purpose large multimodal models. Building upon Gemini's multimodal models, we develop several models within the new Med-Gemini family that inherit core capabilities of Gemini and are optimized for medical use via fine-tuning with 2D and 3D radiology, histopathology, ophthalmology, dermatology and genomic data. Med-Gemini-2D sets a new standard for AI-based chest X-ray (CXR) report generation based on expert evaluation, exceeding previous best results across two separate datasets by an absolute margin of 1% and 12%, where 57% and 96% of AI reports on normal cases, and 43% and 65% on abnormal cases, are evaluated as "equivalent or better" than the original radiologists' reports. We demonstrate the first ever large multimodal model-based report generation for 3D computed tomography (CT) volumes using Med-Gemini-3D, with 53% of AI reports considered clinically acceptable, although additional research is needed to meet expert radiologist reporting quality. Beyond report generation, Med-Gemini-2D surpasses the previous best performance in CXR visual question answering (VQA) and performs well in CXR classification and radiology VQA, exceeding SoTA or baselines on 17 of 20 tasks. In histopathology, ophthalmology, and dermatology image classification, Med-Gemini-2D surpasses baselines across 18 out of 20 tasks and approaches task-specific model performance. Beyond imaging, Med-Gemini-Polygenic outperforms the standard linear polygenic risk score-based approach for disease risk prediction and generalizes to genetically correlated diseases for which it has never been trained. Although further development and evaluation are necessary in the safety-critical medical domain, our results highlight the potential of Med-Gemini across a wide range of medical tasks.
Consensus, dissensus and synergy between clinicians and specialist foundation models in radiology report generation
Dec 06, 2023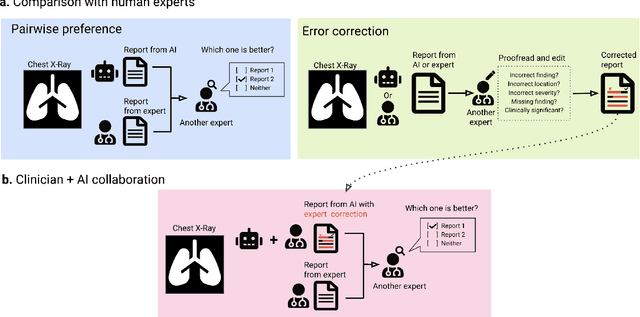
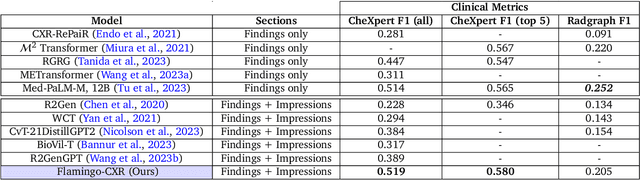
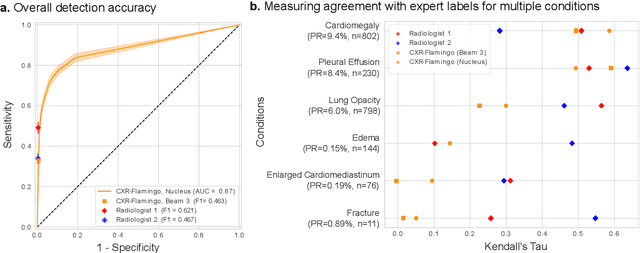
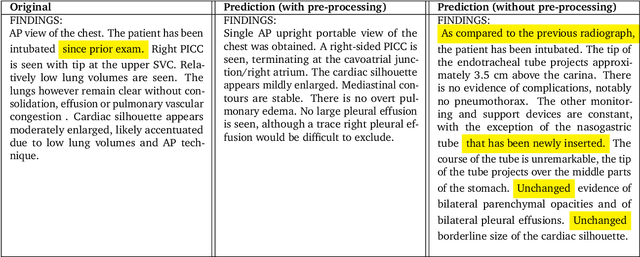
Abstract:Radiology reports are an instrumental part of modern medicine, informing key clinical decisions such as diagnosis and treatment. The worldwide shortage of radiologists, however, restricts access to expert care and imposes heavy workloads, contributing to avoidable errors and delays in report delivery. While recent progress in automated report generation with vision-language models offer clear potential in ameliorating the situation, the path to real-world adoption has been stymied by the challenge of evaluating the clinical quality of AI-generated reports. In this study, we build a state-of-the-art report generation system for chest radiographs, \textit{Flamingo-CXR}, by fine-tuning a well-known vision-language foundation model on radiology data. To evaluate the quality of the AI-generated reports, a group of 16 certified radiologists provide detailed evaluations of AI-generated and human written reports for chest X-rays from an intensive care setting in the United States and an inpatient setting in India. At least one radiologist (out of two per case) preferred the AI report to the ground truth report in over 60$\%$ of cases for both datasets. Amongst the subset of AI-generated reports that contain errors, the most frequently cited reasons were related to the location and finding, whereas for human written reports, most mistakes were related to severity and finding. This disparity suggested potential complementarity between our AI system and human experts, prompting us to develop an assistive scenario in which \textit{Flamingo-CXR} generates a first-draft report, which is subsequently revised by a clinician. This is the first demonstration of clinician-AI collaboration for report writing, and the resultant reports are assessed to be equivalent or preferred by at least one radiologist to reports written by experts alone in 80$\%$ of in-patient cases and 60$\%$ of intensive care cases.
Towards Generalist Biomedical AI
Jul 26, 2023



Abstract:Medicine is inherently multimodal, with rich data modalities spanning text, imaging, genomics, and more. Generalist biomedical artificial intelligence (AI) systems that flexibly encode, integrate, and interpret this data at scale can potentially enable impactful applications ranging from scientific discovery to care delivery. To enable the development of these models, we first curate MultiMedBench, a new multimodal biomedical benchmark. MultiMedBench encompasses 14 diverse tasks such as medical question answering, mammography and dermatology image interpretation, radiology report generation and summarization, and genomic variant calling. We then introduce Med-PaLM Multimodal (Med-PaLM M), our proof of concept for a generalist biomedical AI system. Med-PaLM M is a large multimodal generative model that flexibly encodes and interprets biomedical data including clinical language, imaging, and genomics with the same set of model weights. Med-PaLM M reaches performance competitive with or exceeding the state of the art on all MultiMedBench tasks, often surpassing specialist models by a wide margin. We also report examples of zero-shot generalization to novel medical concepts and tasks, positive transfer learning across tasks, and emergent zero-shot medical reasoning. To further probe the capabilities and limitations of Med-PaLM M, we conduct a radiologist evaluation of model-generated (and human) chest X-ray reports and observe encouraging performance across model scales. In a side-by-side ranking on 246 retrospective chest X-rays, clinicians express a pairwise preference for Med-PaLM M reports over those produced by radiologists in up to 40.50% of cases, suggesting potential clinical utility. While considerable work is needed to validate these models in real-world use cases, our results represent a milestone towards the development of generalist biomedical AI systems.
Constrained Causal Bayesian Optimization
May 31, 2023



Abstract:We propose constrained causal Bayesian optimization (cCBO), an approach for finding interventions in a known causal graph that optimize a target variable under some constraints. cCBO first reduces the search space by exploiting the graph structure and, if available, an observational dataset; and then solves the restricted optimization problem by modelling target and constraint quantities using Gaussian processes and by sequentially selecting interventions via a constrained expected improvement acquisition function. We propose different surrogate models that enable to integrate observational and interventional data while capturing correlation among effects with increasing levels of sophistication. We evaluate cCBO on artificial and real-world causal graphs showing successful trade off between fast convergence and percentage of feasible interventions.
Generative models improve fairness of medical classifiers under distribution shifts
Apr 18, 2023Abstract:A ubiquitous challenge in machine learning is the problem of domain generalisation. This can exacerbate bias against groups or labels that are underrepresented in the datasets used for model development. Model bias can lead to unintended harms, especially in safety-critical applications like healthcare. Furthermore, the challenge is compounded by the difficulty of obtaining labelled data due to high cost or lack of readily available domain expertise. In our work, we show that learning realistic augmentations automatically from data is possible in a label-efficient manner using generative models. In particular, we leverage the higher abundance of unlabelled data to capture the underlying data distribution of different conditions and subgroups for an imaging modality. By conditioning generative models on appropriate labels, we can steer the distribution of synthetic examples according to specific requirements. We demonstrate that these learned augmentations can surpass heuristic ones by making models more robust and statistically fair in- and out-of-distribution. To evaluate the generality of our approach, we study 3 distinct medical imaging contexts of varying difficulty: (i) histopathology images from a publicly available generalisation benchmark, (ii) chest X-rays from publicly available clinical datasets, and (iii) dermatology images characterised by complex shifts and imaging conditions. Complementing real training samples with synthetic ones improves the robustness of models in all three medical tasks and increases fairness by improving the accuracy of diagnosis within underrepresented groups. This approach leads to stark improvements OOD across modalities: 7.7% prediction accuracy improvement in histopathology, 5.2% in chest radiology with 44.6% lower fairness gap and a striking 63.5% improvement in high-risk sensitivity for dermatology with a 7.5x reduction in fairness gap.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge