Ting Chen
ECGFlowCMR: Pretraining with ECG-Generated Cine CMR Improves Cardiac Disease Classification and Phenotype Prediction
Jan 28, 2026Abstract:Cardiac Magnetic Resonance (CMR) imaging provides a comprehensive assessment of cardiac structure and function but remains constrained by high acquisition costs and reliance on expert annotations, limiting the availability of large-scale labeled datasets. In contrast, electrocardiograms (ECGs) are inexpensive, widely accessible, and offer a promising modality for conditioning the generative synthesis of cine CMR. To this end, we propose ECGFlowCMR, a novel ECG-to-CMR generative framework that integrates a Phase-Aware Masked Autoencoder (PA-MAE) and an Anatomy-Motion Disentangled Flow (AMDF) to address two fundamental challenges: (1) the cross-modal temporal mismatch between multi-beat ECG recordings and single-cycle CMR sequences, and (2) the anatomical observability gap due to the limited structural information inherent in ECGs. Extensive experiments on the UK Biobank and a proprietary clinical dataset demonstrate that ECGFlowCMR can generate realistic cine CMR sequences from ECG inputs, enabling scalable pretraining and improving performance on downstream cardiac disease classification and phenotype prediction tasks.
Data Augmentation in Time Series Forecasting through Inverted Framework
Jul 16, 2025Abstract:Currently, iTransformer is one of the most popular and effective models for multivariate time series (MTS) forecasting. Thanks to its inverted framework, iTransformer effectively captures multivariate correlation. However, the inverted framework still has some limitations. It diminishes temporal interdependency information, and introduces noise in cases of nonsignificant variable correlation. To address these limitations, we introduce a novel data augmentation method on inverted framework, called DAIF. Unlike previous data augmentation methods, DAIF stands out as the first real-time augmentation specifically designed for the inverted framework in MTS forecasting. We first define the structure of the inverted sequence-to-sequence framework, then propose two different DAIF strategies, Frequency Filtering and Cross-variation Patching to address the existing challenges of the inverted framework. Experiments across multiple datasets and inverted models have demonstrated the effectiveness of our DAIF.
RareAgents: Autonomous Multi-disciplinary Team for Rare Disease Diagnosis and Treatment
Dec 17, 2024



Abstract:Rare diseases, despite their low individual incidence, collectively impact around 300 million people worldwide due to the huge number of diseases. The complexity of symptoms and the shortage of specialized doctors with relevant experience make diagnosing and treating rare diseases more challenging than common diseases. Recently, agents powered by large language models (LLMs) have demonstrated notable improvements across various domains. In the medical field, some agent methods have outperformed direct prompts in question-answering tasks from medical exams. However, current agent frameworks lack adaptation for real-world clinical scenarios, especially those involving the intricate demands of rare diseases. To address these challenges, we present RareAgents, the first multi-disciplinary team of LLM-based agents tailored to the complex clinical context of rare diseases. RareAgents integrates advanced planning capabilities, memory mechanisms, and medical tools utilization, leveraging Llama-3.1-8B/70B as the base model. Experimental results show that RareAgents surpasses state-of-the-art domain-specific models, GPT-4o, and existing agent frameworks in both differential diagnosis and medication recommendation for rare diseases. Furthermore, we contribute a novel dataset, MIMIC-IV-Ext-Rare, derived from MIMIC-IV, to support further advancements in this field.
Large-scale cross-modality pretrained model enhances cardiovascular state estimation and cardiomyopathy detection from electrocardiograms: An AI system development and multi-center validation study
Nov 19, 2024



Abstract:Cardiovascular diseases (CVDs) present significant challenges for early and accurate diagnosis. While cardiac magnetic resonance imaging (CMR) is the gold standard for assessing cardiac function and diagnosing CVDs, its high cost and technical complexity limit accessibility. In contrast, electrocardiography (ECG) offers promise for large-scale early screening. This study introduces CardiacNets, an innovative model that enhances ECG analysis by leveraging the diagnostic strengths of CMR through cross-modal contrastive learning and generative pretraining. CardiacNets serves two primary functions: (1) it evaluates detailed cardiac function indicators and screens for potential CVDs, including coronary artery disease, cardiomyopathy, pericarditis, heart failure and pulmonary hypertension, using ECG input; and (2) it enhances interpretability by generating high-quality CMR images from ECG data. We train and validate the proposed CardiacNets on two large-scale public datasets (the UK Biobank with 41,519 individuals and the MIMIC-IV-ECG comprising 501,172 samples) as well as three private datasets (FAHZU with 410 individuals, SAHZU with 464 individuals, and QPH with 338 individuals), and the findings demonstrate that CardiacNets consistently outperforms traditional ECG-only models, substantially improving screening accuracy. Furthermore, the generated CMR images provide valuable diagnostic support for physicians of all experience levels. This proof-of-concept study highlights how ECG can facilitate cross-modal insights into cardiac function assessment, paving the way for enhanced CVD screening and diagnosis at a population level.
All-in-one Weather-degraded Image Restoration via Adaptive Degradation-aware Self-prompting Model
Nov 12, 2024



Abstract:Existing approaches for all-in-one weather-degraded image restoration suffer from inefficiencies in leveraging degradation-aware priors, resulting in sub-optimal performance in adapting to different weather conditions. To this end, we develop an adaptive degradation-aware self-prompting model (ADSM) for all-in-one weather-degraded image restoration. Specifically, our model employs the contrastive language-image pre-training model (CLIP) to facilitate the training of our proposed latent prompt generators (LPGs), which represent three types of latent prompts to characterize the degradation type, degradation property and image caption. Moreover, we integrate the acquired degradation-aware prompts into the time embedding of diffusion model to improve degradation perception. Meanwhile, we employ the latent caption prompt to guide the reverse sampling process using the cross-attention mechanism, thereby guiding the accurate image reconstruction. Furthermore, to accelerate the reverse sampling procedure of diffusion model and address the limitations of frequency perception, we introduce a wavelet-oriented noise estimating network (WNE-Net). Extensive experiments conducted on eight publicly available datasets demonstrate the effectiveness of our proposed approach in both task-specific and all-in-one applications.
VividMed: Vision Language Model with Versatile Visual Grounding for Medicine
Oct 16, 2024



Abstract:Recent advancements in Vision Language Models (VLMs) have demonstrated remarkable promise in generating visually grounded responses. However, their application in the medical domain is hindered by unique challenges. For instance, most VLMs rely on a single method of visual grounding, whereas complex medical tasks demand more versatile approaches. Additionally, while most VLMs process only 2D images, a large portion of medical images are 3D. The lack of medical data further compounds these obstacles. To address these challenges, we present VividMed, a vision language model with versatile visual grounding for medicine. Our model supports generating both semantic segmentation masks and instance-level bounding boxes, and accommodates various imaging modalities, including both 2D and 3D data. We design a three-stage training procedure and an automatic data synthesis pipeline based on open datasets and models. Besides visual grounding tasks, VividMed also excels in other common downstream tasks, including Visual Question Answering (VQA) and report generation. Ablation studies empirically show that the integration of visual grounding ability leads to improved performance on these tasks. Our code is publicly available at https://github.com/function2-llx/MMMM.
Unpaired Photo-realistic Image Deraining with Energy-informed Diffusion Model
Jul 24, 2024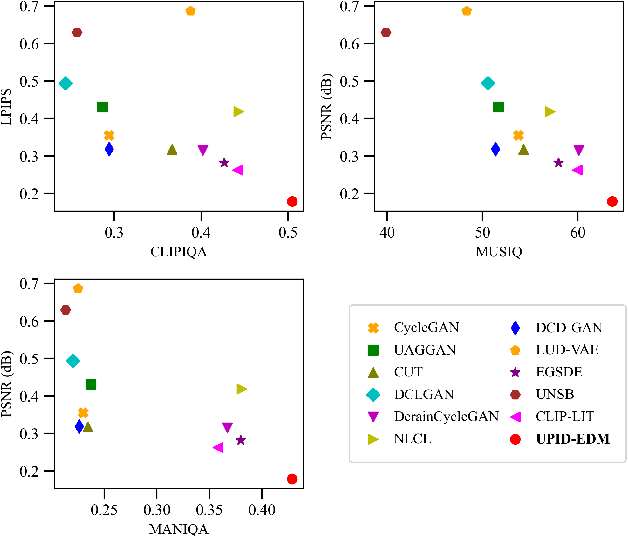
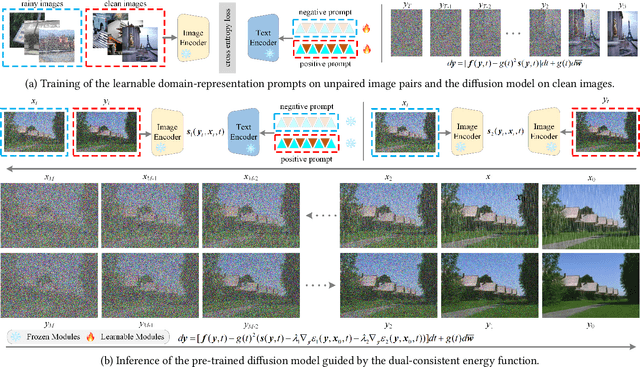
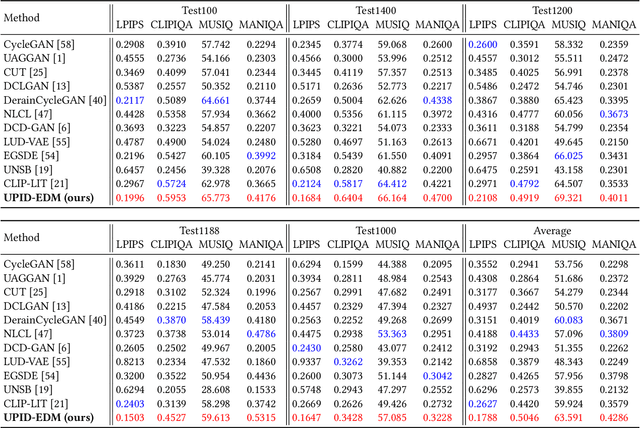
Abstract:Existing unpaired image deraining approaches face challenges in accurately capture the distinguishing characteristics between the rainy and clean domains, resulting in residual degradation and color distortion within the reconstructed images. To this end, we propose an energy-informed diffusion model for unpaired photo-realistic image deraining (UPID-EDM). Initially, we delve into the intricate visual-language priors embedded within the contrastive language-image pre-training model (CLIP), and demonstrate that the CLIP priors aid in the discrimination of rainy and clean images. Furthermore, we introduce a dual-consistent energy function (DEF) that retains the rain-irrelevant characteristics while eliminating the rain-relevant features. This energy function is trained by the non-corresponding rainy and clean images. In addition, we employ the rain-relevance discarding energy function (RDEF) and the rain-irrelevance preserving energy function (RPEF) to direct the reverse sampling procedure of a pre-trained diffusion model, effectively removing the rain streaks while preserving the image contents. Extensive experiments demonstrate that our energy-informed model surpasses the existing unpaired learning approaches in terms of both supervised and no-reference metrics.
Layered Diffusion Model for One-Shot High Resolution Text-to-Image Synthesis
Jul 08, 2024
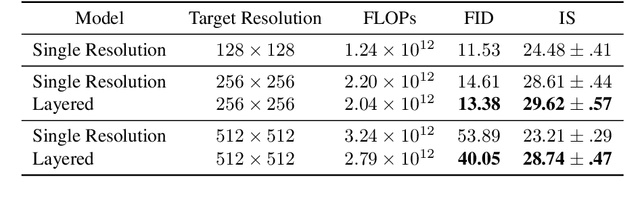


Abstract:We present a one-shot text-to-image diffusion model that can generate high-resolution images from natural language descriptions. Our model employs a layered U-Net architecture that simultaneously synthesizes images at multiple resolution scales. We show that this method outperforms the baseline of synthesizing images only at the target resolution, while reducing the computational cost per step. We demonstrate that higher resolution synthesis can be achieved by layering convolutions at additional resolution scales, in contrast to other methods which require additional models for super-resolution synthesis.
HeMeNet: Heterogeneous Multichannel Equivariant Network for Protein Multitask Learning
Apr 02, 2024



Abstract:Understanding and leveraging the 3D structures of proteins is central to a variety of biological and drug discovery tasks. While deep learning has been applied successfully for structure-based protein function prediction tasks, current methods usually employ distinct training for each task. However, each of the tasks is of small size, and such a single-task strategy hinders the models' performance and generalization ability. As some labeled 3D protein datasets are biologically related, combining multi-source datasets for larger-scale multi-task learning is one way to overcome this problem. In this paper, we propose a neural network model to address multiple tasks jointly upon the input of 3D protein structures. In particular, we first construct a standard structure-based multi-task benchmark called Protein-MT, consisting of 6 biologically relevant tasks, including affinity prediction and property prediction, integrated from 4 public datasets. Then, we develop a novel graph neural network for multi-task learning, dubbed Heterogeneous Multichannel Equivariant Network (HeMeNet), which is E(3) equivariant and able to capture heterogeneous relationships between different atoms. Besides, HeMeNet can achieve task-specific learning via the task-aware readout mechanism. Extensive evaluations on our benchmark verify the effectiveness of multi-task learning, and our model generally surpasses state-of-the-art models.
Denoising Autoregressive Representation Learning
Mar 08, 2024Abstract:In this paper, we explore a new generative approach for learning visual representations. Our method, DARL, employs a decoder-only Transformer to predict image patches autoregressively. We find that training with Mean Squared Error (MSE) alone leads to strong representations. To enhance the image generation ability, we replace the MSE loss with the diffusion objective by using a denoising patch decoder. We show that the learned representation can be improved by using tailored noise schedules and longer training in larger models. Notably, the optimal schedule differs significantly from the typical ones used in standard image diffusion models. Overall, despite its simple architecture, DARL delivers performance remarkably close to state-of-the-art masked prediction models under the fine-tuning protocol. This marks an important step towards a unified model capable of both visual perception and generation, effectively combining the strengths of autoregressive and denoising diffusion models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge