Xiaohao Mao
RareAgents: Autonomous Multi-disciplinary Team for Rare Disease Diagnosis and Treatment
Dec 17, 2024



Abstract:Rare diseases, despite their low individual incidence, collectively impact around 300 million people worldwide due to the huge number of diseases. The complexity of symptoms and the shortage of specialized doctors with relevant experience make diagnosing and treating rare diseases more challenging than common diseases. Recently, agents powered by large language models (LLMs) have demonstrated notable improvements across various domains. In the medical field, some agent methods have outperformed direct prompts in question-answering tasks from medical exams. However, current agent frameworks lack adaptation for real-world clinical scenarios, especially those involving the intricate demands of rare diseases. To address these challenges, we present RareAgents, the first multi-disciplinary team of LLM-based agents tailored to the complex clinical context of rare diseases. RareAgents integrates advanced planning capabilities, memory mechanisms, and medical tools utilization, leveraging Llama-3.1-8B/70B as the base model. Experimental results show that RareAgents surpasses state-of-the-art domain-specific models, GPT-4o, and existing agent frameworks in both differential diagnosis and medication recommendation for rare diseases. Furthermore, we contribute a novel dataset, MIMIC-IV-Ext-Rare, derived from MIMIC-IV, to support further advancements in this field.
RareBench: Can LLMs Serve as Rare Diseases Specialists?
Feb 09, 2024



Abstract:Generalist Large Language Models (LLMs), such as GPT-4, have shown considerable promise in various domains, including medical diagnosis. Rare diseases, affecting approximately 300 million people worldwide, often have unsatisfactory clinical diagnosis rates primarily due to a lack of experienced physicians and the complexity of differentiating among many rare diseases. In this context, recent news such as "ChatGPT correctly diagnosed a 4-year-old's rare disease after 17 doctors failed" underscore LLMs' potential, yet underexplored, role in clinically diagnosing rare diseases. To bridge this research gap, we introduce RareBench, a pioneering benchmark designed to systematically evaluate the capabilities of LLMs on 4 critical dimensions within the realm of rare diseases. Meanwhile, we have compiled the largest open-source dataset on rare disease patients, establishing a benchmark for future studies in this domain. To facilitate differential diagnosis of rare diseases, we develop a dynamic few-shot prompt methodology, leveraging a comprehensive rare disease knowledge graph synthesized from multiple knowledge bases, significantly enhancing LLMs' diagnostic performance. Moreover, we present an exhaustive comparative study of GPT-4's diagnostic capabilities against those of specialist physicians. Our experimental findings underscore the promising potential of integrating LLMs into the clinical diagnostic process for rare diseases. This paves the way for exciting possibilities in future advancements in this field.
FIT: a Fast and Accurate Framework for Solving Medical Inquiring and Diagnosing Tasks
Dec 02, 2020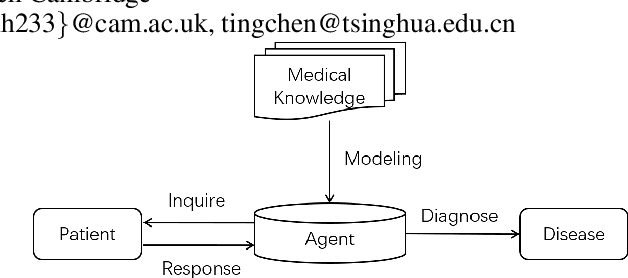
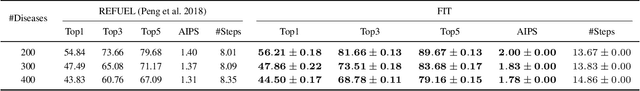
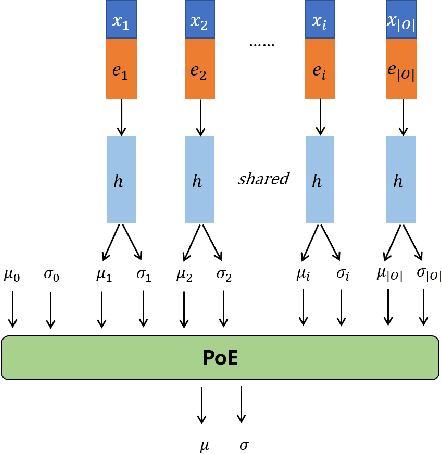
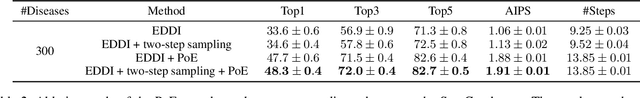
Abstract:Automatic self-diagnosis provides low-cost and accessible healthcare via an agent that queries the patient and makes predictions about possible diseases. From a machine learning perspective, symptom-based self-diagnosis can be viewed as a sequential feature selection and classification problem. Reinforcement learning methods have shown good performance in this task but often suffer from large search spaces and costly training. To address these problems, we propose a competitive framework, called FIT, which uses an information-theoretic reward to determine what data to collect next. FIT improves over previous information-based approaches by using a multimodal variational autoencoder (MVAE) model and a two-step sampling strategy for disease prediction. Furthermore, we propose novel methods to substantially reduce the computational cost of FIT to a level that is acceptable for practical online self-diagnosis. Our results in two simulated datasets show that FIT can effectively deal with large search space problems, outperforming existing baselines. Moreover, using two medical datasets, we show that FIT is a competitive alternative in real-world settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge