Xuanzhong Chen
BabyVision: Visual Reasoning Beyond Language
Jan 10, 2026Abstract:While humans develop core visual skills long before acquiring language, contemporary Multimodal LLMs (MLLMs) still rely heavily on linguistic priors to compensate for their fragile visual understanding. We uncovered a crucial fact: state-of-the-art MLLMs consistently fail on basic visual tasks that humans, even 3-year-olds, can solve effortlessly. To systematically investigate this gap, we introduce BabyVision, a benchmark designed to assess core visual abilities independent of linguistic knowledge for MLLMs. BabyVision spans a wide range of tasks, with 388 items divided into 22 subclasses across four key categories. Empirical results and human evaluation reveal that leading MLLMs perform significantly below human baselines. Gemini3-Pro-Preview scores 49.7, lagging behind 6-year-old humans and falling well behind the average adult score of 94.1. These results show despite excelling in knowledge-heavy evaluations, current MLLMs still lack fundamental visual primitives. Progress in BabyVision represents a step toward human-level visual perception and reasoning capabilities. We also explore solving visual reasoning with generation models by proposing BabyVision-Gen and automatic evaluation toolkit. Our code and benchmark data are released at https://github.com/UniPat-AI/BabyVision for reproduction.
EcomBench: Towards Holistic Evaluation of Foundation Agents in E-commerce
Dec 11, 2025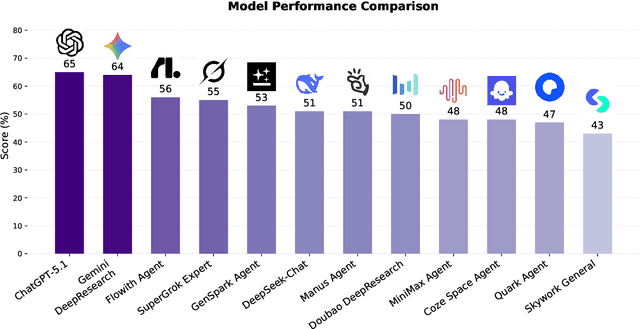
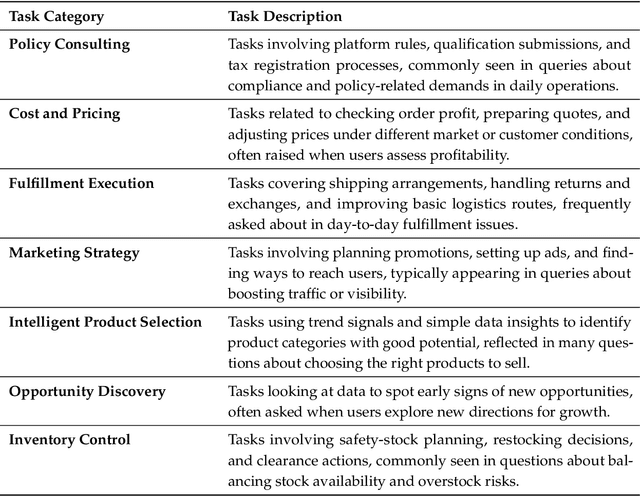

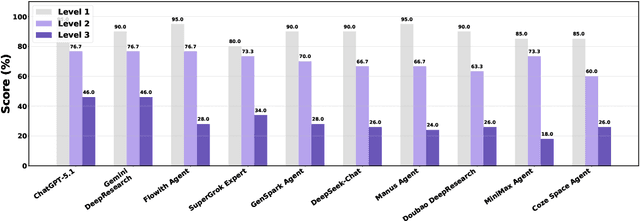
Abstract:Foundation agents have rapidly advanced in their ability to reason and interact with real environments, making the evaluation of their core capabilities increasingly important. While many benchmarks have been developed to assess agent performance, most concentrate on academic settings or artificially designed scenarios while overlooking the challenges that arise in real applications. To address this issue, we focus on a highly practical real-world setting, the e-commerce domain, which involves a large volume of diverse user interactions, dynamic market conditions, and tasks directly tied to real decision-making processes. To this end, we introduce EcomBench, a holistic E-commerce Benchmark designed to evaluate agent performance in realistic e-commerce environments. EcomBench is built from genuine user demands embedded in leading global e-commerce ecosystems and is carefully curated and annotated through human experts to ensure clarity, accuracy, and domain relevance. It covers multiple task categories within e-commerce scenarios and defines three difficulty levels that evaluate agents on key capabilities such as deep information retrieval, multi-step reasoning, and cross-source knowledge integration. By grounding evaluation in real e-commerce contexts, EcomBench provides a rigorous and dynamic testbed for measuring the practical capabilities of agents in modern e-commerce.
IterResearch: Rethinking Long-Horizon Agents via Markovian State Reconstruction
Nov 10, 2025Abstract:Recent advances in deep-research agents have shown promise for autonomous knowledge construction through dynamic reasoning over external sources. However, existing approaches rely on a mono-contextual paradigm that accumulates all information in a single, expanding context window, leading to context suffocation and noise contamination that limit their effectiveness on long-horizon tasks. We introduce IterResearch, a novel iterative deep-research paradigm that reformulates long-horizon research as a Markov Decision Process with strategic workspace reconstruction. By maintaining an evolving report as memory and periodically synthesizing insights, our approach preserves consistent reasoning capacity across arbitrary exploration depths. We further develop Efficiency-Aware Policy Optimization (EAPO), a reinforcement learning framework that incentivizes efficient exploration through geometric reward discounting and enables stable distributed training via adaptive downsampling. Extensive experiments demonstrate that IterResearch achieves substantial improvements over existing open-source agents with average +14.5pp across six benchmarks and narrows the gap with frontier proprietary systems. Remarkably, our paradigm exhibits unprecedented interaction scaling, extending to 2048 interactions with dramatic performance gains (from 3.5\% to 42.5\%), and serves as an effective prompting strategy, improving frontier models by up to 19.2pp over ReAct on long-horizon tasks. These findings position IterResearch as a versatile solution for long-horizon reasoning, effective both as a trained agent and as a prompting paradigm for frontier models.
MARS: Optimizing Dual-System Deep Research via Multi-Agent Reinforcement Learning
Oct 06, 2025Abstract:Large Reasoning Models (LRMs) often exhibit a tendency for overanalysis in simple tasks, where the models excessively utilize System 2-type, deliberate reasoning, leading to inefficient token generation. Furthermore, these models face challenges in adapting their reasoning capabilities to rapidly changing environments due to the static nature of their pretraining data. To address these issues, advancing Large Language Models (LLMs) for complex reasoning tasks requires innovative approaches that bridge intuitive and deliberate cognitive processes, akin to human cognition's dual-system dynamic. This paper introduces a Multi-Agent System for Deep ReSearch (MARS) enabling seamless integration of System 1's fast, intuitive thinking with System 2's deliberate reasoning within LLMs. MARS strategically integrates multiple external tools, such as Google Search, Google Scholar, and Python Interpreter, to access up-to-date information and execute complex computations, while creating a specialized division of labor where System 1 efficiently processes and summarizes high-volume external information, providing distilled insights that expand System 2's reasoning context without overwhelming its capacity. Furthermore, we propose a multi-agent reinforcement learning framework extending Group Relative Policy Optimization to simultaneously optimize both systems with multi-turn tool interactions, bin-packing optimization, and sample balancing strategies that enhance collaborative efficiency. Extensive experiments demonstrate MARS achieves substantial improvements of 3.86% on the challenging Humanity's Last Exam (HLE) benchmark and an average gain of 8.9% across 7 knowledge-intensive tasks, validating the effectiveness of our dual-system paradigm for complex reasoning in dynamic information environments.
Scaling Agents via Continual Pre-training
Sep 16, 2025Abstract:Large language models (LLMs) have evolved into agentic systems capable of autonomous tool use and multi-step reasoning for complex problem-solving. However, post-training approaches building upon general-purpose foundation models consistently underperform in agentic tasks, particularly in open-source implementations. We identify the root cause: the absence of robust agentic foundation models forces models during post-training to simultaneously learn diverse agentic behaviors while aligning them to expert demonstrations, thereby creating fundamental optimization tensions. To this end, we are the first to propose incorporating Agentic Continual Pre-training (Agentic CPT) into the deep research agents training pipeline to build powerful agentic foundational models. Based on this approach, we develop a deep research agent model named AgentFounder. We evaluate our AgentFounder-30B on 10 benchmarks and achieve state-of-the-art performance while retains strong tool-use ability, notably 39.9% on BrowseComp-en, 43.3% on BrowseComp-zh, and 31.5% Pass@1 on HLE.
WebResearcher: Unleashing unbounded reasoning capability in Long-Horizon Agents
Sep 16, 2025



Abstract:Recent advances in deep-research systems have demonstrated the potential for AI agents to autonomously discover and synthesize knowledge from external sources. In this paper, we introduce WebResearcher, a novel framework for building such agents through two key components: (1) WebResearcher, an iterative deep-research paradigm that reformulates deep research as a Markov Decision Process, where agents periodically consolidate findings into evolving reports while maintaining focused workspaces, overcoming the context suffocation and noise contamination that plague existing mono-contextual approaches; and (2) WebFrontier, a scalable data synthesis engine that generates high-quality training data through tool-augmented complexity escalation, enabling systematic creation of research tasks that bridge the gap between passive knowledge recall and active knowledge construction. Notably, we find that the training data from our paradigm significantly enhances tool-use capabilities even for traditional mono-contextual methods. Furthermore, our paradigm naturally scales through parallel thinking, enabling concurrent multi-agent exploration for more comprehensive conclusions. Extensive experiments across 6 challenging benchmarks demonstrate that WebResearcher achieves state-of-the-art performance, even surpassing frontier proprietary systems.
RareAgents: Autonomous Multi-disciplinary Team for Rare Disease Diagnosis and Treatment
Dec 17, 2024



Abstract:Rare diseases, despite their low individual incidence, collectively impact around 300 million people worldwide due to the huge number of diseases. The complexity of symptoms and the shortage of specialized doctors with relevant experience make diagnosing and treating rare diseases more challenging than common diseases. Recently, agents powered by large language models (LLMs) have demonstrated notable improvements across various domains. In the medical field, some agent methods have outperformed direct prompts in question-answering tasks from medical exams. However, current agent frameworks lack adaptation for real-world clinical scenarios, especially those involving the intricate demands of rare diseases. To address these challenges, we present RareAgents, the first multi-disciplinary team of LLM-based agents tailored to the complex clinical context of rare diseases. RareAgents integrates advanced planning capabilities, memory mechanisms, and medical tools utilization, leveraging Llama-3.1-8B/70B as the base model. Experimental results show that RareAgents surpasses state-of-the-art domain-specific models, GPT-4o, and existing agent frameworks in both differential diagnosis and medication recommendation for rare diseases. Furthermore, we contribute a novel dataset, MIMIC-IV-Ext-Rare, derived from MIMIC-IV, to support further advancements in this field.
VividMed: Vision Language Model with Versatile Visual Grounding for Medicine
Oct 16, 2024



Abstract:Recent advancements in Vision Language Models (VLMs) have demonstrated remarkable promise in generating visually grounded responses. However, their application in the medical domain is hindered by unique challenges. For instance, most VLMs rely on a single method of visual grounding, whereas complex medical tasks demand more versatile approaches. Additionally, while most VLMs process only 2D images, a large portion of medical images are 3D. The lack of medical data further compounds these obstacles. To address these challenges, we present VividMed, a vision language model with versatile visual grounding for medicine. Our model supports generating both semantic segmentation masks and instance-level bounding boxes, and accommodates various imaging modalities, including both 2D and 3D data. We design a three-stage training procedure and an automatic data synthesis pipeline based on open datasets and models. Besides visual grounding tasks, VividMed also excels in other common downstream tasks, including Visual Question Answering (VQA) and report generation. Ablation studies empirically show that the integration of visual grounding ability leads to improved performance on these tasks. Our code is publicly available at https://github.com/function2-llx/MMMM.
RareBench: Can LLMs Serve as Rare Diseases Specialists?
Feb 09, 2024



Abstract:Generalist Large Language Models (LLMs), such as GPT-4, have shown considerable promise in various domains, including medical diagnosis. Rare diseases, affecting approximately 300 million people worldwide, often have unsatisfactory clinical diagnosis rates primarily due to a lack of experienced physicians and the complexity of differentiating among many rare diseases. In this context, recent news such as "ChatGPT correctly diagnosed a 4-year-old's rare disease after 17 doctors failed" underscore LLMs' potential, yet underexplored, role in clinically diagnosing rare diseases. To bridge this research gap, we introduce RareBench, a pioneering benchmark designed to systematically evaluate the capabilities of LLMs on 4 critical dimensions within the realm of rare diseases. Meanwhile, we have compiled the largest open-source dataset on rare disease patients, establishing a benchmark for future studies in this domain. To facilitate differential diagnosis of rare diseases, we develop a dynamic few-shot prompt methodology, leveraging a comprehensive rare disease knowledge graph synthesized from multiple knowledge bases, significantly enhancing LLMs' diagnostic performance. Moreover, we present an exhaustive comparative study of GPT-4's diagnostic capabilities against those of specialist physicians. Our experimental findings underscore the promising potential of integrating LLMs into the clinical diagnostic process for rare diseases. This paves the way for exciting possibilities in future advancements in this field.
Pre-trained Universal Medical Image Transformer
Dec 12, 2023Abstract:Self-supervised learning has emerged as a viable method to leverage the abundance of unlabeled medical imaging data, addressing the challenge of labeled data scarcity in medical image analysis. In particular, masked image modeling (MIM) with visual token reconstruction has shown promising results in the general computer vision (CV) domain and serves as a candidate for medical image analysis. However, the presence of heterogeneous 2D and 3D medical images often limits the volume and diversity of training data that can be effectively used for a single model structure. In this work, we propose a spatially adaptive convolution (SAC) module, which adaptively adjusts convolution parameters based on the voxel spacing of the input images. Employing this SAC module, we build a universal visual tokenizer and a universal Vision Transformer (ViT) capable of effectively processing a wide range of medical images with various imaging modalities and spatial properties. Moreover, in order to enhance the robustness of the visual tokenizer's reconstruction objective for MIM, we suggest to generalize the discrete token output of the visual tokenizer to a probabilistic soft token. We show that the generalized soft token representation can be effectively integrated with the prior distribution regularization through a constructive interpretation. As a result, we pre-train a universal visual tokenizer followed by a universal ViT via visual token reconstruction on 55 public medical image datasets, comprising over 9 million 2D slices (including over 48,000 3D images). This represents the largest, most comprehensive, and diverse dataset for pre-training 3D medical image models to our knowledge. Experimental results on downstream medical image classification and segmentation tasks demonstrate the superior performance of our model and improved label efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge