Lingxiao Luo
EJ
VividMed: Vision Language Model with Versatile Visual Grounding for Medicine
Oct 16, 2024



Abstract:Recent advancements in Vision Language Models (VLMs) have demonstrated remarkable promise in generating visually grounded responses. However, their application in the medical domain is hindered by unique challenges. For instance, most VLMs rely on a single method of visual grounding, whereas complex medical tasks demand more versatile approaches. Additionally, while most VLMs process only 2D images, a large portion of medical images are 3D. The lack of medical data further compounds these obstacles. To address these challenges, we present VividMed, a vision language model with versatile visual grounding for medicine. Our model supports generating both semantic segmentation masks and instance-level bounding boxes, and accommodates various imaging modalities, including both 2D and 3D data. We design a three-stage training procedure and an automatic data synthesis pipeline based on open datasets and models. Besides visual grounding tasks, VividMed also excels in other common downstream tasks, including Visual Question Answering (VQA) and report generation. Ablation studies empirically show that the integration of visual grounding ability leads to improved performance on these tasks. Our code is publicly available at https://github.com/function2-llx/MMMM.
HeMeNet: Heterogeneous Multichannel Equivariant Network for Protein Multitask Learning
Apr 02, 2024



Abstract:Understanding and leveraging the 3D structures of proteins is central to a variety of biological and drug discovery tasks. While deep learning has been applied successfully for structure-based protein function prediction tasks, current methods usually employ distinct training for each task. However, each of the tasks is of small size, and such a single-task strategy hinders the models' performance and generalization ability. As some labeled 3D protein datasets are biologically related, combining multi-source datasets for larger-scale multi-task learning is one way to overcome this problem. In this paper, we propose a neural network model to address multiple tasks jointly upon the input of 3D protein structures. In particular, we first construct a standard structure-based multi-task benchmark called Protein-MT, consisting of 6 biologically relevant tasks, including affinity prediction and property prediction, integrated from 4 public datasets. Then, we develop a novel graph neural network for multi-task learning, dubbed Heterogeneous Multichannel Equivariant Network (HeMeNet), which is E(3) equivariant and able to capture heterogeneous relationships between different atoms. Besides, HeMeNet can achieve task-specific learning via the task-aware readout mechanism. Extensive evaluations on our benchmark verify the effectiveness of multi-task learning, and our model generally surpasses state-of-the-art models.
Pre-trained Universal Medical Image Transformer
Dec 12, 2023Abstract:Self-supervised learning has emerged as a viable method to leverage the abundance of unlabeled medical imaging data, addressing the challenge of labeled data scarcity in medical image analysis. In particular, masked image modeling (MIM) with visual token reconstruction has shown promising results in the general computer vision (CV) domain and serves as a candidate for medical image analysis. However, the presence of heterogeneous 2D and 3D medical images often limits the volume and diversity of training data that can be effectively used for a single model structure. In this work, we propose a spatially adaptive convolution (SAC) module, which adaptively adjusts convolution parameters based on the voxel spacing of the input images. Employing this SAC module, we build a universal visual tokenizer and a universal Vision Transformer (ViT) capable of effectively processing a wide range of medical images with various imaging modalities and spatial properties. Moreover, in order to enhance the robustness of the visual tokenizer's reconstruction objective for MIM, we suggest to generalize the discrete token output of the visual tokenizer to a probabilistic soft token. We show that the generalized soft token representation can be effectively integrated with the prior distribution regularization through a constructive interpretation. As a result, we pre-train a universal visual tokenizer followed by a universal ViT via visual token reconstruction on 55 public medical image datasets, comprising over 9 million 2D slices (including over 48,000 3D images). This represents the largest, most comprehensive, and diverse dataset for pre-training 3D medical image models to our knowledge. Experimental results on downstream medical image classification and segmentation tasks demonstrate the superior performance of our model and improved label efficiency.
Overview of the Ninth Dialog System Technology Challenge: DSTC9
Nov 12, 2020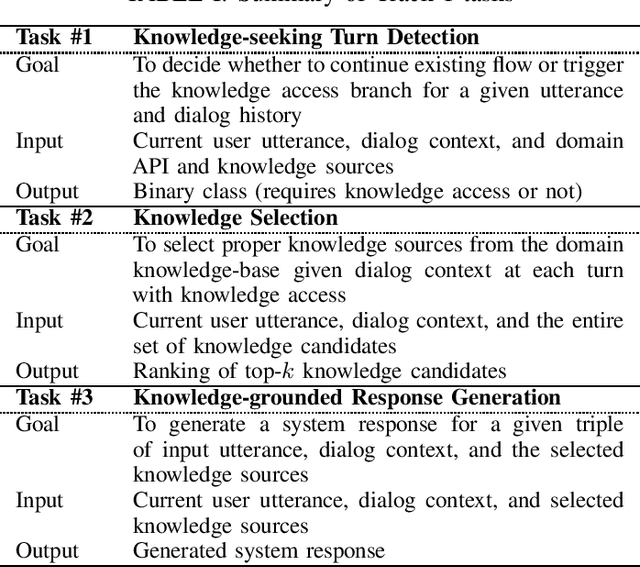
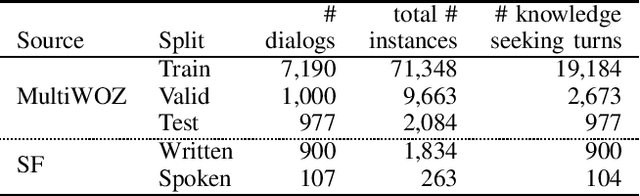

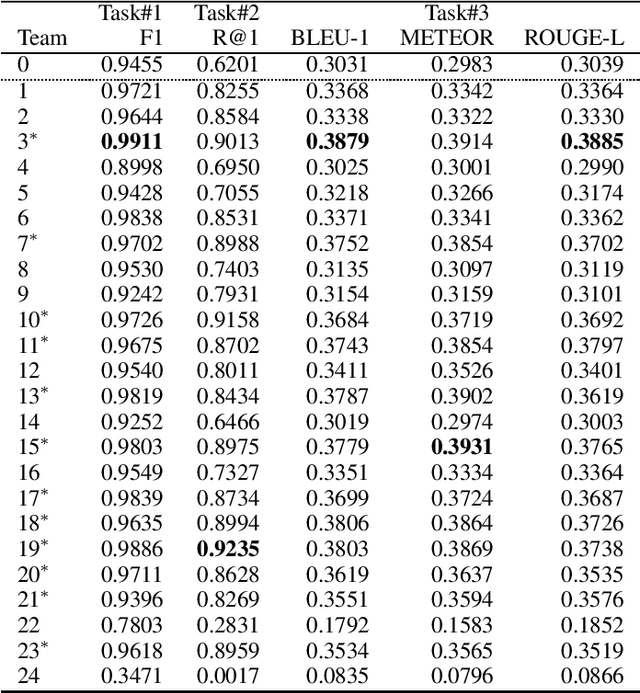
Abstract:This paper introduces the Ninth Dialog System Technology Challenge (DSTC-9). This edition of the DSTC focuses on applying end-to-end dialog technologies for four distinct tasks in dialog systems, namely, 1. Task-oriented dialog Modeling with unstructured knowledge access, 2. Multi-domain task-oriented dialog, 3. Interactive evaluation of dialog, and 4. Situated interactive multi-modal dialog. This paper describes the task definition, provided datasets, baselines and evaluation set-up for each track. We also summarize the results of the submitted systems to highlight the overall trends of the state-of-the-art technologies for the tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge