Ziyu Li
High-fidelity 3D multi-slab diffusion MRI using Slab-shifting for Harmonized 3D Acquisition and Reconstruction with Profile Encoding Networks (SHARPEN)
Feb 06, 2026Abstract:Three-dimensional (3D) multi-slab imaging is a promising approach for high-resolution in vivo diffusion MRI (dMRI) due to its compatibility with short TR (1-2 s), providing optimal signal-to-noise ratio (SNR) efficiency. A major challenge, however, is slab boundary artifacts arising from non-ideal slab-selective RF excitation. Non-rectangular slab profiles reduce signal intensity at slab boundaries, while profile overlap across adjacent slabs introduces inter-slab crosstalk, where repeated excitation shortens the local TR and limits T1 recovery. To mitigate slab boundary artifacts without increasing scan time, we build on slab profile encoding and propose Slab-shifting for Harmonized 3D Acquisition and Reconstruction with Profile Encoding Networks (SHARPEN). For different diffusion directions, SHARPEN applies inter-volume field-of-view shifts along the slice direction to provide complementary slab profile encoding without prolonging acquisition. Slab profiles are estimated using a lightweight self-supervised neural network that exploits consistency across shifted acquisitions and known physical properties of slab profiles and diffusion images, and corrected images are reconstructed accordingly. SHARPEN was validated using simulated and prospectively acquired high-resolution in vivo data and demonstrates accurate slab profile estimation and robust boundary artifact correction, even in the presence of inter-volume motion. SHARPEN does not require high-quality reference training data and supports subject-specific training. Its efficient GPU-based implementation delivers faster and more accurate correction than NPEN, yielding slice-wise quantitative profiles that closely match those from reference 2D acquisitions. SHARPEN enables high-quality dMRI at 0.7 mm isotropic resolution on a 3T clinical scanner, highlighting its potential to advance submillimeter dMRI for neuroscience research.
From Prompt to Graph: Comparing LLM-Based Information Extraction Strategies in Domain-Specific Ontology Development
Jan 31, 2026Abstract:Ontologies are essential for structuring domain knowledge, improving accessibility, sharing, and reuse. However, traditional ontology construction relies on manual annotation and conventional natural language processing (NLP) techniques, making the process labour-intensive and costly, especially in specialised fields like casting manufacturing. The rise of Large Language Models (LLMs) offers new possibilities for automating knowledge extraction. This study investigates three LLM-based approaches, including pre-trained LLM-driven method, in-context learning (ICL) method and fine-tuning method to extract terms and relations from domain-specific texts using limited data. We compare their performances and use the best-performing method to build a casting ontology that validated by domian expert.
Parallel Diffusion Solver via Residual Dirichlet Policy Optimization
Dec 28, 2025Abstract:Diffusion models (DMs) have achieved state-of-the-art generative performance but suffer from high sampling latency due to their sequential denoising nature. Existing solver-based acceleration methods often face significant image quality degradation under a low-latency budget, primarily due to accumulated truncation errors arising from the inability to capture high-curvature trajectory segments. In this paper, we propose the Ensemble Parallel Direction solver (dubbed as EPD-Solver), a novel ODE solver that mitigates these errors by incorporating multiple parallel gradient evaluations in each step. Motivated by the geometric insight that sampling trajectories are largely confined to a low-dimensional manifold, EPD-Solver leverages the Mean Value Theorem for vector-valued functions to approximate the integral solution more accurately. Importantly, since the additional gradient computations are independent, they can be fully parallelized, preserving low-latency sampling nature. We introduce a two-stage optimization framework. Initially, EPD-Solver optimizes a small set of learnable parameters via a distillation-based approach. We further propose a parameter-efficient Reinforcement Learning (RL) fine-tuning scheme that reformulates the solver as a stochastic Dirichlet policy. Unlike traditional methods that fine-tune the massive backbone, our RL approach operates strictly within the low-dimensional solver space, effectively mitigating reward hacking while enhancing performance in complex text-to-image (T2I) generation tasks. In addition, our method is flexible and can serve as a plugin (EPD-Plugin) to improve existing ODE samplers.
NeuroCLIP: Brain-Inspired Prompt Tuning for EEG-to-Image Multimodal Contrastive Learning
Nov 12, 2025Abstract:Recent advances in brain-inspired artificial intelligence have sought to align neural signals with visual semantics using multimodal models such as CLIP. However, existing methods often treat CLIP as a static feature extractor, overlooking its adaptability to neural representations and the inherent physiological-symbolic gap in EEG-image alignment. To address these challenges, we present NeuroCLIP, a prompt tuning framework tailored for EEG-to-image contrastive learning. Our approach introduces three core innovations: (1) We design a dual-stream visual embedding pipeline that combines dynamic filtering and token-level fusion to generate instance-level adaptive prompts, which guide the adjustment of patch embedding tokens based on image content, thereby enabling fine-grained modulation of visual representations under neural constraints; (2) We are the first to introduce visual prompt tokens into EEG-image alignment, acting as global, modality-level prompts that work in conjunction with instance-level adjustments. These visual prompt tokens are inserted into the Transformer architecture to facilitate neural-aware adaptation and parameter optimization at a global level; (3) Inspired by neuroscientific principles of human visual encoding, we propose a refined contrastive loss that better model the semantic ambiguity and cross-modal noise present in EEG signals. On the THINGS-EEG2 dataset, NeuroCLIP achieves a Top-1 accuracy of 63.2% in zero-shot image retrieval, surpassing the previous best method by +12.3%, and demonstrates strong generalization under inter-subject conditions (+4.6% Top-1), highlighting the potential of physiology-aware prompt tuning for bridging brain signals and visual semantics.
DC-Seg: Disentangled Contrastive Learning for Brain Tumor Segmentation with Missing Modalities
May 17, 2025Abstract:Accurate segmentation of brain images typically requires the integration of complementary information from multiple image modalities. However, clinical data for all modalities may not be available for every patient, creating a significant challenge. To address this, previous studies encode multiple modalities into a shared latent space. While somewhat effective, it remains suboptimal, as each modality contains distinct and valuable information. In this study, we propose DC-Seg (Disentangled Contrastive Learning for Segmentation), a new method that explicitly disentangles images into modality-invariant anatomical representation and modality-specific representation, by using anatomical contrastive learning and modality contrastive learning respectively. This solution improves the separation of anatomical and modality-specific features by considering the modality gaps, leading to more robust representations. Furthermore, we introduce a segmentation-based regularizer that enhances the model's robustness to missing modalities. Extensive experiments on the BraTS 2020 and a private white matter hyperintensity(WMH) segmentation dataset demonstrate that DC-Seg outperforms state-of-the-art methods in handling incomplete multimodal brain tumor segmentation tasks with varying missing modalities, while also demonstrate strong generalizability in WMH segmentation. The code is available at https://github.com/CuCl-2/DC-Seg.
Phenotype-Guided Generative Model for High-Fidelity Cardiac MRI Synthesis: Advancing Pretraining and Clinical Applications
May 06, 2025



Abstract:Cardiac Magnetic Resonance (CMR) imaging is a vital non-invasive tool for diagnosing heart diseases and evaluating cardiac health. However, the limited availability of large-scale, high-quality CMR datasets poses a major challenge to the effective application of artificial intelligence (AI) in this domain. Even the amount of unlabeled data and the health status it covers are difficult to meet the needs of model pretraining, which hinders the performance of AI models on downstream tasks. In this study, we present Cardiac Phenotype-Guided CMR Generation (CPGG), a novel approach for generating diverse CMR data that covers a wide spectrum of cardiac health status. The CPGG framework consists of two stages: in the first stage, a generative model is trained using cardiac phenotypes derived from CMR data; in the second stage, a masked autoregressive diffusion model, conditioned on these phenotypes, generates high-fidelity CMR cine sequences that capture both structural and functional features of the heart in a fine-grained manner. We synthesized a massive amount of CMR to expand the pretraining data. Experimental results show that CPGG generates high-quality synthetic CMR data, significantly improving performance on various downstream tasks, including diagnosis and cardiac phenotypes prediction. These gains are demonstrated across both public and private datasets, highlighting the effectiveness of our approach. Code is availabel at https://anonymous.4open.science/r/CPGG.
Brain Inspired Adaptive Memory Dual-Net for Few-Shot Image Classification
Mar 10, 2025Abstract:Few-shot image classification has become a popular research topic for its wide application in real-world scenarios, however the problem of supervision collapse induced by single image-level annotation remains a major challenge. Existing methods aim to tackle this problem by locating and aligning relevant local features. However, the high intra-class variability in real-world images poses significant challenges in locating semantically relevant local regions under few-shot settings. Drawing inspiration from the human's complementary learning system, which excels at rapidly capturing and integrating semantic features from limited examples, we propose the generalization-optimized Systems Consolidation Adaptive Memory Dual-Network, SCAM-Net. This approach simulates the systems consolidation of complementary learning system with an adaptive memory module, which successfully addresses the difficulty of identifying meaningful features in few-shot scenarios. Specifically, we construct a Hippocampus-Neocortex dual-network that consolidates structured representation of each category, the structured representation is then stored and adaptively regulated following the generalization optimization principle in a long-term memory inside Neocortex. Extensive experiments on benchmark datasets show that the proposed model has achieved state-of-the-art performance.
De-biased Multimodal Electrocardiogram Analysis
Nov 22, 2024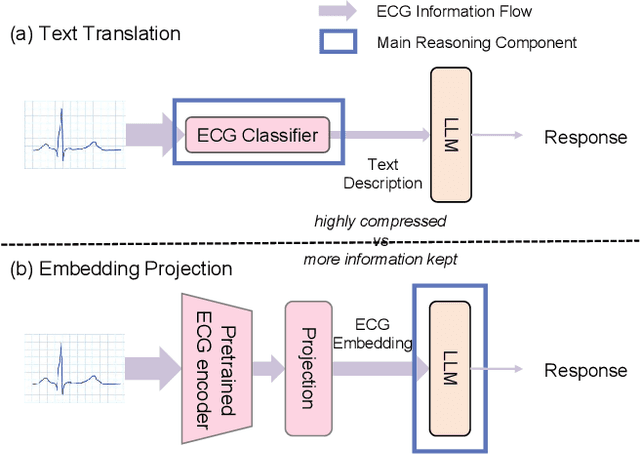
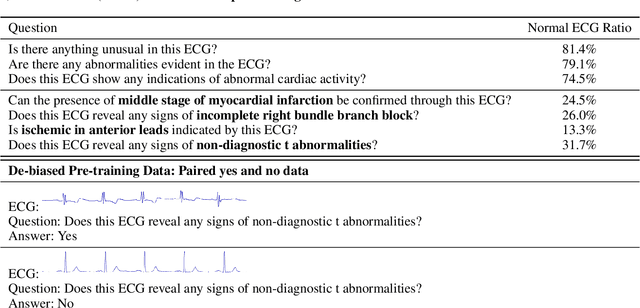
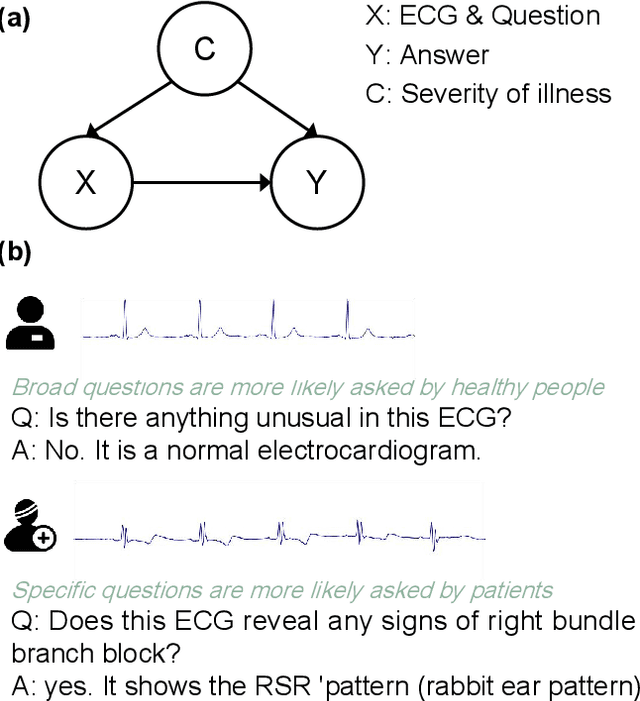

Abstract:Multimodal large language models (MLLMs) are increasingly being applied in the medical field, particularly in medical imaging. However, developing MLLMs for ECG signals, which are crucial in clinical settings, has been a significant challenge beyond medical imaging. Previous studies have attempted to address this by converting ECGs into several text tags using an external classifier in a training-free manner. However, this approach significantly compresses the information in ECGs and underutilizes the reasoning capabilities of LLMs. In this work, we directly feed the embeddings of ECGs into the LLM through a projection layer, retaining more information about ECGs and better leveraging the reasoning abilities of LLMs. Our method can also effectively handle a common situation in clinical practice where it is necessary to compare two ECGs taken at different times. Recent studies found that MLLMs may rely solely on text input to provide answers, ignoring inputs from other modalities. We analyzed this phenomenon from a causal perspective in the context of ECG MLLMs and discovered that the confounder, severity of illness, introduces a spurious correlation between the question and answer, leading the model to rely on this spurious correlation and ignore the ECG input. Such models do not comprehend the ECG input and perform poorly in adversarial tests where different expressions of the same question are used in the training and testing sets. We designed a de-biased pre-training method to eliminate the confounder's effect according to the theory of backdoor adjustment. Our model performed well on the ECG-QA task under adversarial testing and demonstrated zero-shot capabilities. An interesting random ECG test further validated that our model effectively understands and utilizes the input ECG signal.
Large-scale cross-modality pretrained model enhances cardiovascular state estimation and cardiomyopathy detection from electrocardiograms: An AI system development and multi-center validation study
Nov 19, 2024



Abstract:Cardiovascular diseases (CVDs) present significant challenges for early and accurate diagnosis. While cardiac magnetic resonance imaging (CMR) is the gold standard for assessing cardiac function and diagnosing CVDs, its high cost and technical complexity limit accessibility. In contrast, electrocardiography (ECG) offers promise for large-scale early screening. This study introduces CardiacNets, an innovative model that enhances ECG analysis by leveraging the diagnostic strengths of CMR through cross-modal contrastive learning and generative pretraining. CardiacNets serves two primary functions: (1) it evaluates detailed cardiac function indicators and screens for potential CVDs, including coronary artery disease, cardiomyopathy, pericarditis, heart failure and pulmonary hypertension, using ECG input; and (2) it enhances interpretability by generating high-quality CMR images from ECG data. We train and validate the proposed CardiacNets on two large-scale public datasets (the UK Biobank with 41,519 individuals and the MIMIC-IV-ECG comprising 501,172 samples) as well as three private datasets (FAHZU with 410 individuals, SAHZU with 464 individuals, and QPH with 338 individuals), and the findings demonstrate that CardiacNets consistently outperforms traditional ECG-only models, substantially improving screening accuracy. Furthermore, the generated CMR images provide valuable diagnostic support for physicians of all experience levels. This proof-of-concept study highlights how ECG can facilitate cross-modal insights into cardiac function assessment, paving the way for enhanced CVD screening and diagnosis at a population level.
LLM-PQA: LLM-enhanced Prediction Query Answering
Sep 02, 2024



Abstract:The advent of Large Language Models (LLMs) provides an opportunity to change the way queries are processed, moving beyond the constraints of conventional SQL-based database systems. However, using an LLM to answer a prediction query is still challenging, since an external ML model has to be employed and inference has to be performed in order to provide an answer. This paper introduces LLM-PQA, a novel tool that addresses prediction queries formulated in natural language. LLM-PQA is the first to combine the capabilities of LLMs and retrieval-augmented mechanism for the needs of prediction queries by integrating data lakes and model zoos. This integration provides users with access to a vast spectrum of heterogeneous data and diverse ML models, facilitating dynamic prediction query answering. In addition, LLM-PQA can dynamically train models on demand, based on specific query requirements, ensuring reliable and relevant results even when no pre-trained model in a model zoo, available for the task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge