Yiheng Mao
DC-Seg: Disentangled Contrastive Learning for Brain Tumor Segmentation with Missing Modalities
May 17, 2025Abstract:Accurate segmentation of brain images typically requires the integration of complementary information from multiple image modalities. However, clinical data for all modalities may not be available for every patient, creating a significant challenge. To address this, previous studies encode multiple modalities into a shared latent space. While somewhat effective, it remains suboptimal, as each modality contains distinct and valuable information. In this study, we propose DC-Seg (Disentangled Contrastive Learning for Segmentation), a new method that explicitly disentangles images into modality-invariant anatomical representation and modality-specific representation, by using anatomical contrastive learning and modality contrastive learning respectively. This solution improves the separation of anatomical and modality-specific features by considering the modality gaps, leading to more robust representations. Furthermore, we introduce a segmentation-based regularizer that enhances the model's robustness to missing modalities. Extensive experiments on the BraTS 2020 and a private white matter hyperintensity(WMH) segmentation dataset demonstrate that DC-Seg outperforms state-of-the-art methods in handling incomplete multimodal brain tumor segmentation tasks with varying missing modalities, while also demonstrate strong generalizability in WMH segmentation. The code is available at https://github.com/CuCl-2/DC-Seg.
Phenotype-Guided Generative Model for High-Fidelity Cardiac MRI Synthesis: Advancing Pretraining and Clinical Applications
May 06, 2025



Abstract:Cardiac Magnetic Resonance (CMR) imaging is a vital non-invasive tool for diagnosing heart diseases and evaluating cardiac health. However, the limited availability of large-scale, high-quality CMR datasets poses a major challenge to the effective application of artificial intelligence (AI) in this domain. Even the amount of unlabeled data and the health status it covers are difficult to meet the needs of model pretraining, which hinders the performance of AI models on downstream tasks. In this study, we present Cardiac Phenotype-Guided CMR Generation (CPGG), a novel approach for generating diverse CMR data that covers a wide spectrum of cardiac health status. The CPGG framework consists of two stages: in the first stage, a generative model is trained using cardiac phenotypes derived from CMR data; in the second stage, a masked autoregressive diffusion model, conditioned on these phenotypes, generates high-fidelity CMR cine sequences that capture both structural and functional features of the heart in a fine-grained manner. We synthesized a massive amount of CMR to expand the pretraining data. Experimental results show that CPGG generates high-quality synthetic CMR data, significantly improving performance on various downstream tasks, including diagnosis and cardiac phenotypes prediction. These gains are demonstrated across both public and private datasets, highlighting the effectiveness of our approach. Code is availabel at https://anonymous.4open.science/r/CPGG.
De-biased Multimodal Electrocardiogram Analysis
Nov 22, 2024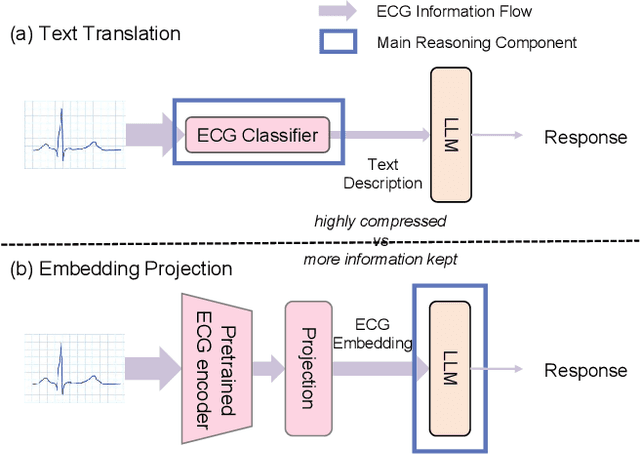
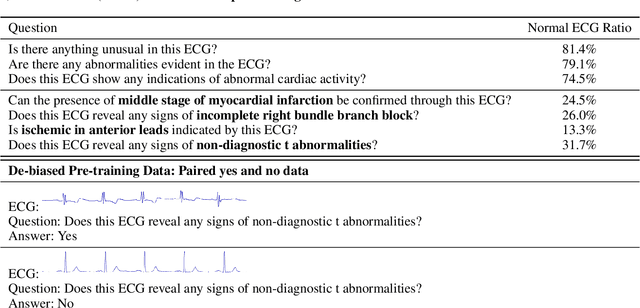
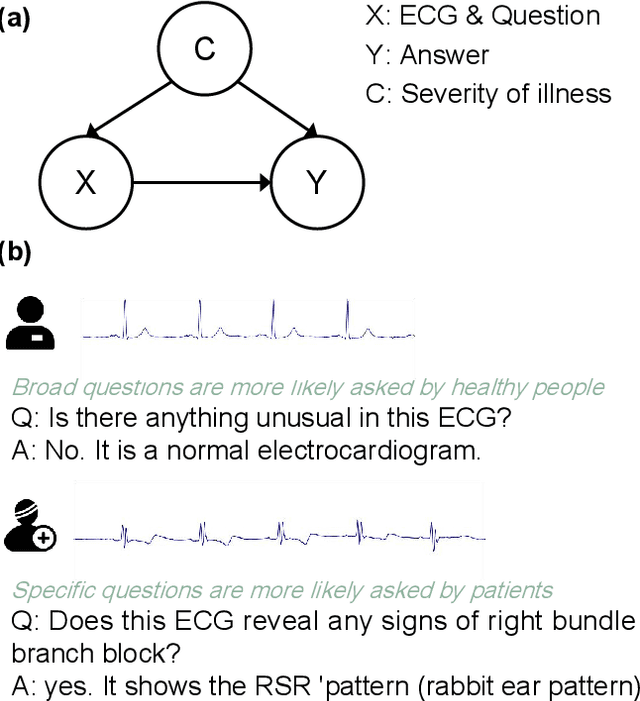

Abstract:Multimodal large language models (MLLMs) are increasingly being applied in the medical field, particularly in medical imaging. However, developing MLLMs for ECG signals, which are crucial in clinical settings, has been a significant challenge beyond medical imaging. Previous studies have attempted to address this by converting ECGs into several text tags using an external classifier in a training-free manner. However, this approach significantly compresses the information in ECGs and underutilizes the reasoning capabilities of LLMs. In this work, we directly feed the embeddings of ECGs into the LLM through a projection layer, retaining more information about ECGs and better leveraging the reasoning abilities of LLMs. Our method can also effectively handle a common situation in clinical practice where it is necessary to compare two ECGs taken at different times. Recent studies found that MLLMs may rely solely on text input to provide answers, ignoring inputs from other modalities. We analyzed this phenomenon from a causal perspective in the context of ECG MLLMs and discovered that the confounder, severity of illness, introduces a spurious correlation between the question and answer, leading the model to rely on this spurious correlation and ignore the ECG input. Such models do not comprehend the ECG input and perform poorly in adversarial tests where different expressions of the same question are used in the training and testing sets. We designed a de-biased pre-training method to eliminate the confounder's effect according to the theory of backdoor adjustment. Our model performed well on the ECG-QA task under adversarial testing and demonstrated zero-shot capabilities. An interesting random ECG test further validated that our model effectively understands and utilizes the input ECG signal.
Large-scale cross-modality pretrained model enhances cardiovascular state estimation and cardiomyopathy detection from electrocardiograms: An AI system development and multi-center validation study
Nov 19, 2024



Abstract:Cardiovascular diseases (CVDs) present significant challenges for early and accurate diagnosis. While cardiac magnetic resonance imaging (CMR) is the gold standard for assessing cardiac function and diagnosing CVDs, its high cost and technical complexity limit accessibility. In contrast, electrocardiography (ECG) offers promise for large-scale early screening. This study introduces CardiacNets, an innovative model that enhances ECG analysis by leveraging the diagnostic strengths of CMR through cross-modal contrastive learning and generative pretraining. CardiacNets serves two primary functions: (1) it evaluates detailed cardiac function indicators and screens for potential CVDs, including coronary artery disease, cardiomyopathy, pericarditis, heart failure and pulmonary hypertension, using ECG input; and (2) it enhances interpretability by generating high-quality CMR images from ECG data. We train and validate the proposed CardiacNets on two large-scale public datasets (the UK Biobank with 41,519 individuals and the MIMIC-IV-ECG comprising 501,172 samples) as well as three private datasets (FAHZU with 410 individuals, SAHZU with 464 individuals, and QPH with 338 individuals), and the findings demonstrate that CardiacNets consistently outperforms traditional ECG-only models, substantially improving screening accuracy. Furthermore, the generated CMR images provide valuable diagnostic support for physicians of all experience levels. This proof-of-concept study highlights how ECG can facilitate cross-modal insights into cardiac function assessment, paving the way for enhanced CVD screening and diagnosis at a population level.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge