Ari Ercole
Generalised Label-free Artefact Cleaning for Real-time Medical Pulsatile Time Series
Apr 29, 2025Abstract:Artefacts compromise clinical decision-making in the use of medical time series. Pulsatile waveforms offer probabilities for accurate artefact detection, yet most approaches rely on supervised manners and overlook patient-level distribution shifts. To address these issues, we introduce a generalised label-free framework, GenClean, for real-time artefact cleaning and leverage an in-house dataset of 180,000 ten-second arterial blood pressure (ABP) samples for training. We first investigate patient-level generalisation, demonstrating robust performances under both intra- and inter-patient distribution shifts. We further validate its effectiveness through challenging cross-disease cohort experiments on the MIMIC-III database. Additionally, we extend our method to photoplethysmography (PPG), highlighting its applicability to diverse medical pulsatile signals. Finally, its integration into ICM+, a clinical research monitoring software, confirms the real-time feasibility of our framework, emphasising its practical utility in continuous physiological monitoring. This work provides a foundational step toward precision medicine in improving the reliability of high-resolution medical time series analysis
Clairvoyance: A Pipeline Toolkit for Medical Time Series
Oct 28, 2023Abstract:Time-series learning is the bread and butter of data-driven *clinical decision support*, and the recent explosion in ML research has demonstrated great potential in various healthcare settings. At the same time, medical time-series problems in the wild are challenging due to their highly *composite* nature: They entail design choices and interactions among components that preprocess data, impute missing values, select features, issue predictions, estimate uncertainty, and interpret models. Despite exponential growth in electronic patient data, there is a remarkable gap between the potential and realized utilization of ML for clinical research and decision support. In particular, orchestrating a real-world project lifecycle poses challenges in engineering (i.e. hard to build), evaluation (i.e. hard to assess), and efficiency (i.e. hard to optimize). Designed to address these issues simultaneously, Clairvoyance proposes a unified, end-to-end, autoML-friendly pipeline that serves as a (i) software toolkit, (ii) empirical standard, and (iii) interface for optimization. Our ultimate goal lies in facilitating transparent and reproducible experimentation with complex inference workflows, providing integrated pathways for (1) personalized prediction, (2) treatment-effect estimation, and (3) information acquisition. Through illustrative examples on real-world data in outpatient, general wards, and intensive-care settings, we illustrate the applicability of the pipeline paradigm on core tasks in the healthcare journey. To the best of our knowledge, Clairvoyance is the first to demonstrate viability of a comprehensive and automatable pipeline for clinical time-series ML.
Contribution of clinical course to outcome after traumatic brain injury: mining patient trajectories from European intensive care unit data
Mar 08, 2023Abstract:Existing methods to characterise the evolving condition of traumatic brain injury (TBI) patients in the intensive care unit (ICU) do not capture the context necessary for individualising treatment. We aimed to develop a modelling strategy which integrates all data stored in medical records to produce an interpretable disease course for each TBI patient's ICU stay. From a prospective, European cohort (n=1,550, 65 centres, 19 countries) of TBI patients, we extracted all 1,166 variables collected before or during ICU stay as well as 6-month functional outcome on the Glasgow Outcome Scale-Extended (GOSE). We trained recurrent neural network models to map a token-embedded time series representation of all variables (including missing data) to an ordinal GOSE prognosis every 2 hours. With repeated cross-validation, we evaluated calibration and the explanation of ordinal variance in GOSE with Somers' Dxy. Furthermore, we applied TimeSHAP to calculate the contribution of variables and prior timepoints towards transitions in patient trajectories. Our modelling strategy achieved calibration at 8 hours, and the full range of variables explained up to 52% (95% CI: 50-54%) of the variance in ordinal functional outcome. Up to 91% (90-91%) of this explanation was derived from pre-ICU and admission information. Information collected in the ICU increased explanation (by up to 5% [4-6%]), though not enough to counter poorer performance in longer-stay (>5.75 days) patients. Static variables with the highest contributions were physician prognoses and certain demographic and CT features. Among dynamic variables, markers of intracranial hypertension and neurological function contributed the most. Whilst static information currently accounts for the majority of functional outcome explanation, our data-driven analysis highlights investigative avenues to improve dynamic characterisation of longer-stay patients.
The leap to ordinal: functional prognosis after traumatic brain injury using artificial intelligence
Feb 10, 2022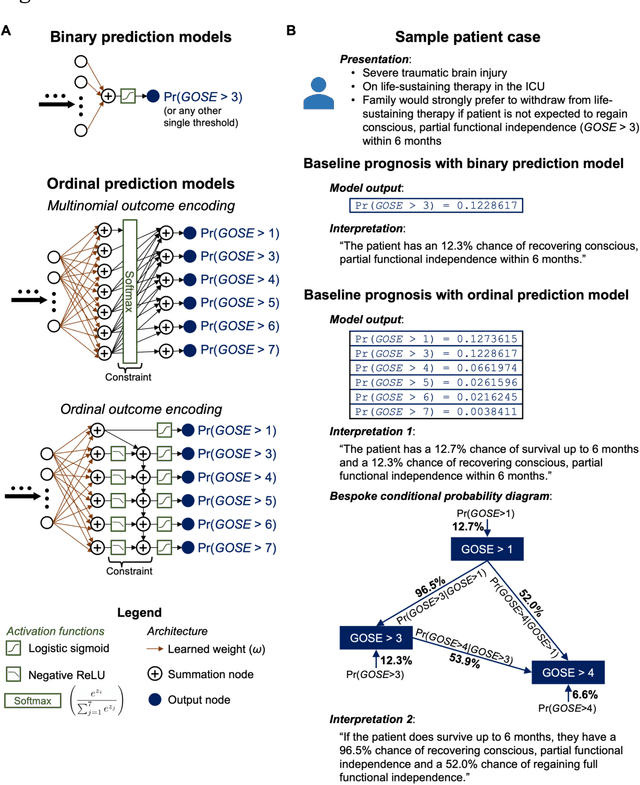
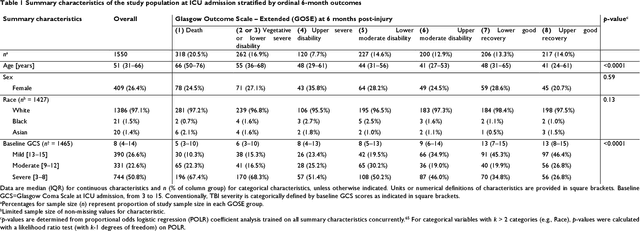
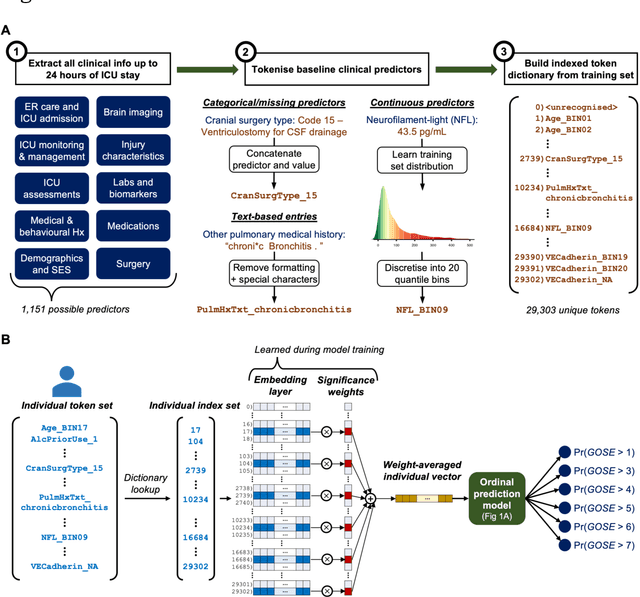
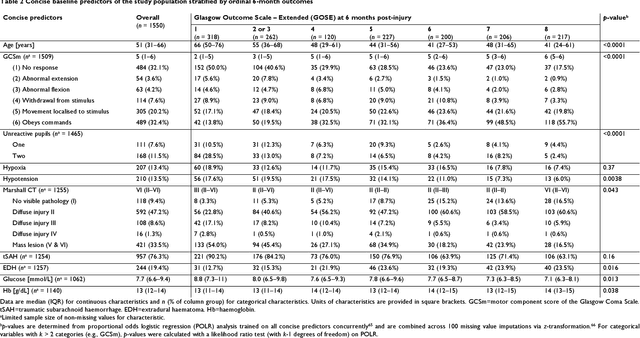
Abstract:When a patient is admitted to the intensive care unit (ICU) after a traumatic brain injury (TBI), an early prognosis is essential for baseline risk adjustment and shared decision making. TBI outcomes are commonly categorised by the Glasgow Outcome Scale-Extended (GOSE) into 8, ordered levels of functional recovery at 6 months after injury. Existing ICU prognostic models predict binary outcomes at a certain threshold of GOSE (e.g., prediction of survival [GOSE>1] or functional independence [GOSE>4]). We aimed to develop ordinal prediction models that concurrently predict probabilities of each GOSE score. From a prospective cohort (n=1,550, 65 centres) in the ICU stratum of the Collaborative European NeuroTrauma Effectiveness Research in TBI (CENTER-TBI) patient dataset, we extracted all clinical information within 24 hours of ICU admission (1,151 predictors) and 6-month GOSE scores. We analysed the effect of 2 design elements on ordinal model performance: (1) the baseline predictor set, ranging from a concise set of 10 validated predictors to a token-embedded representation of all possible predictors, and (2) the modelling strategy, from ordinal logistic regression to multinomial deep learning. With repeated k-fold cross-validation, we found that expanding the baseline predictor set significantly improved ordinal prediction performance while increasing analytical complexity did not. Half of these gains could be achieved with the addition of 8 high-impact predictors (2 demographic variables, 4 protein biomarkers, and 2 severity assessments) to the concise set. At best, ordinal models achieved 0.76 (95% CI: 0.74-0.77) ordinal discrimination ability (ordinal c-index) and 57% (95% CI: 54%-60%) explanation of ordinal variation in 6-month GOSE (Somers' D). Our results motivate the search for informative predictors for higher GOSE and the development of ordinal dynamic prediction models.
Hide-and-Seek Privacy Challenge
Jul 24, 2020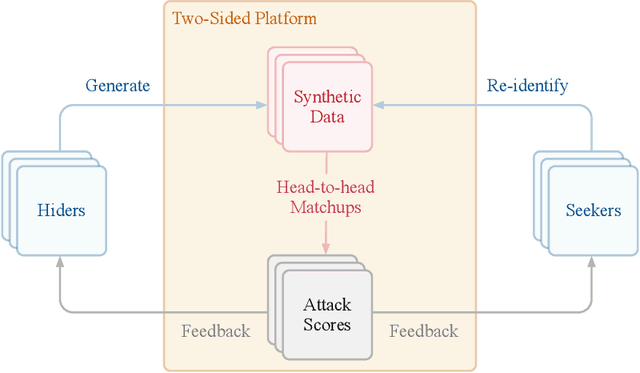
Abstract:The clinical time-series setting poses a unique combination of challenges to data modeling and sharing. Due to the high dimensionality of clinical time series, adequate de-identification to preserve privacy while retaining data utility is difficult to achieve using common de-identification techniques. An innovative approach to this problem is synthetic data generation. From a technical perspective, a good generative model for time-series data should preserve temporal dynamics, in the sense that new sequences respect the original relationships between high-dimensional variables across time. From the privacy perspective, the model should prevent patient re-identification by limiting vulnerability to membership inference attacks. The NeurIPS 2020 Hide-and-Seek Privacy Challenge is a novel two-tracked competition to simultaneously accelerate progress in tackling both problems. In our head-to-head format, participants in the synthetic data generation track (i.e. "hiders") and the patient re-identification track (i.e. "seekers") are directly pitted against each other by way of a new, high-quality intensive care time-series dataset: the AmsterdamUMCdb dataset. Ultimately, we seek to advance generative techniques for dense and high-dimensional temporal data streams that are (1) clinically meaningful in terms of fidelity and predictivity, as well as (2) capable of minimizing membership privacy risks in terms of the concrete notion of patient re-identification.
Adaptive Prediction Timing for Electronic Health Records
Mar 05, 2020



Abstract:In realistic scenarios, multivariate timeseries evolve over case-by-case time-scales. This is particularly clear in medicine, where the rate of clinical events varies by ward, patient, and application. Increasingly complex models have been shown to effectively predict patient outcomes, but have failed to adapt granularity to these inherent temporal resolutions. As such, we introduce a novel, more realistic, approach to generating patient outcome predictions at an adaptive rate based on uncertainty accumulation in Bayesian recurrent models. We use a Recurrent Neural Network (RNN) and a Bayesian embedding layer with a new aggregation method to demonstrate adaptive prediction timing. Our model predicts more frequently when events are dense or the model is certain of event latent representations, and less frequently when readings are sparse or the model is uncertain. At 48 hours after patient admission, our model achieves equal performance compared to its static-windowed counterparts, while generating patient- and event-specific prediction timings that lead to improved predictive performance over the crucial first 12 hours of the patient stay.
Impact of novel aggregation methods for flexible, time-sensitive EHR prediction without variable selection or cleaning
Sep 17, 2019



Abstract:Dynamic assessment of patient status (e.g. by an automated, continuously updated assessment of outcome) in the Intensive Care Unit (ICU) is of paramount importance for early alerting, decision support and resource allocation. Extraction and cleaning of expert-selected clinical variables discards information and protracts collaborative efforts to introduce machine learning in medicine. We present improved aggregation methods for a flexible deep learning architecture which learns a joint representation of patient chart, lab and output events. Our models outperform recent deep learning models for patient mortality classification using ICU timeseries, by embedding and aggregating all events with no pre-processing or variable selection. Our model achieves a strong performance of AUROC 0.87 at 48 hours on the MIMIC-III dataset while using 13,233 unique un-preprocessed variables in an interpretable manner via hourly softmax aggregation. This demonstrates how our method can be easily combined with existing electronic health record systems for automated, dynamic patient risk analysis.
Dynamic survival prediction in intensive care units from heterogeneous time series without the need for variable selection or pre-processing
Sep 17, 2019



Abstract:We present a machine learning pipeline and model that uses the entire uncurated EHR for prediction of in-hospital mortality at arbitrary time intervals, using all available chart, lab and output events, without the need for pre-processing or feature engineering. Data for more than 45,000 American ICU patients from the MIMIC-III database were used to develop an ICU mortality prediction model. All chart, lab and output events were treated by the model in the same manner inspired by Natural Language Processing (NLP). Patient events were discretized by percentile and mapped to learnt embeddings before being passed to a Recurrent Neural Network (RNN) to provide early prediction of in-patient mortality risk. We compared mortality predictions with the Simplified Acute Physiology Score II (SAPS II) and the Oxford Acute Severity of Illness Score (OASIS). Data were split into an independent test set (10%) and a ten-fold cross-validation was carried out during training to avoid overfitting. 13,233 distinct variables with heterogeneous data types were included without manual selection or pre-processing. Recordings in the first few hours of a patient's stay were found to be strongly predictive of mortality, outperforming models using SAPS II and OASIS scores within just 2 hours and achieving a state of the art Area Under the Receiver Operating Characteristic (AUROC) value of 0.80 (95% CI 0.79-0.80) at 12 hours vs 0.70 and 0.66 for SAPS II and OASIS at 24 hours respectively. Our model achieves a very strong performance of AUROC 0.86 (95% CI 0.85-0.86) for in-patient mortality prediction after 48 hours on the MIMIC-III dataset. Predictive performance increases over the first 48 hours of the ICU stay, but suffers from diminishing returns, providing rationale for time-limited trials of critical care and suggesting that the timing of decision making can be optimised and individualised.
DeepClean -- self-supervised artefact rejection for intensive care waveform data using generative deep learning
Sep 05, 2019



Abstract:Waveform physiological data is important in the treatment of critically ill patients in the intensive care unit. Such recordings are susceptible to artefacts, which must be removed before the data can be re-used for alerting or reprocessed for other clinical or research purposes. Accurate removal of artefacts reduces both bias and uncertainty in clinical assessment and the false positive rate of intensive care unit alarms, and is therefore a key component in providing optimal clinical care. In this work, we present DeepClean; a prototype self-supervised artefact detection system using a convolutional variational autoencoder deep neural network that avoids costly and painstaking manual annotation, requiring only easily-obtained 'good' data for training. For a test case with invasive arterial blood pressure, we demonstrate that our algorithm can detect the presence of an artefact within a 10-second sample of data with sensitivity and specificity around 90%. Furthermore, DeepClean was able to identify regions of artefact within such samples with high accuracy and we show that it significantly outperforms a baseline principle component analysis approach in both signal reconstruction and artefact detection. DeepClean learns a generative model and therefore may also be used for imputation of missing data.
Interpretable Outcome Prediction with Sparse Bayesian Neural Networks in Intensive Care
May 07, 2019



Abstract:Clinical decision making is challenging because of pathological complexity, as well as large amounts of heterogeneous data generated as part of routine clinical care. In recent years, machine learning tools have been developed to aid this process. Intensive care unit (ICU) admissions represent the most data dense and time-critical patient care episodes. In this context, prediction models may help clinicians determine which patients are most at risk and prioritize care. However, flexible tools such as artificial neural networks (ANNs) suffer from a lack of interpretability limiting their acceptability to clinicians. In this work, we propose a novel interpretable Bayesian neural network architecture which offers both the flexibility of ANNs and interpretability in terms of feature selection. In particular, we employ a sparsity inducing prior distribution in a tied manner to learn which features are important for outcome prediction. We evaluate our approach on the task of mortality prediction using two real-world ICU cohorts. In collaboration with clinicians we found that, in addition to the predicted outcome results, our approach can provide novel insights into the importance of different clinical measurements. This suggests that our model can support medical experts in their decision making process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge