Jacob Deasy
Fine-tuning Protein Language Models with Deep Mutational Scanning improves Variant Effect Prediction
May 10, 2024
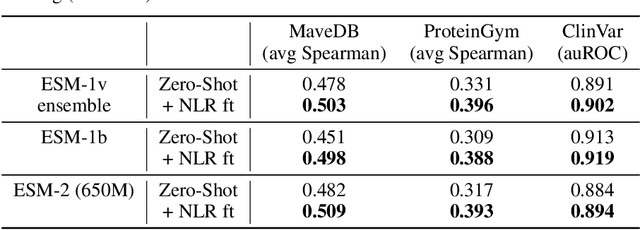

Abstract:Protein Language Models (PLMs) have emerged as performant and scalable tools for predicting the functional impact and clinical significance of protein-coding variants, but they still lag experimental accuracy. Here, we present a novel fine-tuning approach to improve the performance of PLMs with experimental maps of variant effects from Deep Mutational Scanning (DMS) assays using a Normalised Log-odds Ratio (NLR) head. We find consistent improvements in a held-out protein test set, and on independent DMS and clinical variant annotation benchmarks from ProteinGym and ClinVar. These findings demonstrate that DMS is a promising source of sequence diversity and supervised training data for improving the performance of PLMs for variant effect prediction.
Generalising sequence models for epigenome predictions with tissue and assay embeddings
Aug 22, 2023Abstract:Sequence modelling approaches for epigenetic profile prediction have recently expanded in terms of sequence length, model size, and profile diversity. However, current models cannot infer on many experimentally feasible tissue and assay pairs due to poor usage of contextual information, limiting $\textit{in silico}$ understanding of regulatory genomics. We demonstrate that strong correlation can be achieved across a large range of experimental conditions by integrating tissue and assay embeddings into a Contextualised Genomic Network (CGN). In contrast to previous approaches, we enhance long-range sequence embeddings with contextual information in the input space, rather than expanding the output space. We exhibit the efficacy of our approach across a broad set of epigenetic profiles and provide the first insights into the effect of genetic variants on epigenetic sequence model training. Our general approach to context integration exceeds state of the art in multiple settings while employing a more rigorous validation procedure.
Heavy-tailed denoising score matching
Dec 17, 2021



Abstract:Score-based model research in the last few years has produced state of the art generative models by employing Gaussian denoising score-matching (DSM). However, the Gaussian noise assumption has several high-dimensional limitations, motivating a more concrete route toward even higher dimension PDF estimation in future. We outline this limitation, before extending the theory to a broader family of noising distributions -- namely, the generalised normal distribution. To theoretically ground this, we relax a key assumption in (denoising) score matching theory, demonstrating that distributions which are differentiable \textit{almost everywhere} permit the same objective simplification as Gaussians. For noise vector length distributions, we demonstrate favourable concentration of measure in the high-dimensional spaces prevalent in deep learning. In the process, we uncover a skewed noise vector length distribution and develop an iterative noise scaling algorithm to consistently initialise the multiple levels of noise in annealed Langevin dynamics. On the practical side, our use of heavy-tailed DSM leads to improved score estimation, controllable sampling convergence, and more balanced unconditional generative performance for imbalanced datasets.
Constraining Variational Inference with Geometric Jensen-Shannon Divergence
Jun 18, 2020



Abstract:We examine the problem of controlling divergences for latent space regularisation in variational autoencoders. Specifically, when aiming to reconstruct example $x\in\mathbb{R}^{m}$ via latent space $z\in\mathbb{R}^{n}$ ($n\leq m$), while balancing this against the need for generalisable latent representations. We present a regularisation mechanism based on the skew geometric-Jensen-Shannon divergence $\left(\textrm{JS}^{\textrm{G}_{\alpha}}\right)$. We find a variation in $\textrm{JS}^{\textrm{G}_{\alpha}}$, motivated by limiting cases, which leads to an intuitive interpolation between forward and reverse KL in the space of both distributions and divergences. We motivate its potential benefits for VAEs through low-dimensional examples, before presenting quantitative and qualitative results. Our experiments demonstrate that skewing our variant of $\textrm{JS}^{\textrm{G}_{\alpha}}$, in the context of $\textrm{JS}^{\textrm{G}_{\alpha}}$-VAEs, leads to better reconstruction and generation when compared to several baseline VAEs. Our approach is entirely unsupervised and utilises only one hyperparameter which can be easily interpreted in latent space.
Adaptive Prediction Timing for Electronic Health Records
Mar 05, 2020



Abstract:In realistic scenarios, multivariate timeseries evolve over case-by-case time-scales. This is particularly clear in medicine, where the rate of clinical events varies by ward, patient, and application. Increasingly complex models have been shown to effectively predict patient outcomes, but have failed to adapt granularity to these inherent temporal resolutions. As such, we introduce a novel, more realistic, approach to generating patient outcome predictions at an adaptive rate based on uncertainty accumulation in Bayesian recurrent models. We use a Recurrent Neural Network (RNN) and a Bayesian embedding layer with a new aggregation method to demonstrate adaptive prediction timing. Our model predicts more frequently when events are dense or the model is certain of event latent representations, and less frequently when readings are sparse or the model is uncertain. At 48 hours after patient admission, our model achieves equal performance compared to its static-windowed counterparts, while generating patient- and event-specific prediction timings that lead to improved predictive performance over the crucial first 12 hours of the patient stay.
Impact of novel aggregation methods for flexible, time-sensitive EHR prediction without variable selection or cleaning
Sep 17, 2019



Abstract:Dynamic assessment of patient status (e.g. by an automated, continuously updated assessment of outcome) in the Intensive Care Unit (ICU) is of paramount importance for early alerting, decision support and resource allocation. Extraction and cleaning of expert-selected clinical variables discards information and protracts collaborative efforts to introduce machine learning in medicine. We present improved aggregation methods for a flexible deep learning architecture which learns a joint representation of patient chart, lab and output events. Our models outperform recent deep learning models for patient mortality classification using ICU timeseries, by embedding and aggregating all events with no pre-processing or variable selection. Our model achieves a strong performance of AUROC 0.87 at 48 hours on the MIMIC-III dataset while using 13,233 unique un-preprocessed variables in an interpretable manner via hourly softmax aggregation. This demonstrates how our method can be easily combined with existing electronic health record systems for automated, dynamic patient risk analysis.
Dynamic survival prediction in intensive care units from heterogeneous time series without the need for variable selection or pre-processing
Sep 17, 2019



Abstract:We present a machine learning pipeline and model that uses the entire uncurated EHR for prediction of in-hospital mortality at arbitrary time intervals, using all available chart, lab and output events, without the need for pre-processing or feature engineering. Data for more than 45,000 American ICU patients from the MIMIC-III database were used to develop an ICU mortality prediction model. All chart, lab and output events were treated by the model in the same manner inspired by Natural Language Processing (NLP). Patient events were discretized by percentile and mapped to learnt embeddings before being passed to a Recurrent Neural Network (RNN) to provide early prediction of in-patient mortality risk. We compared mortality predictions with the Simplified Acute Physiology Score II (SAPS II) and the Oxford Acute Severity of Illness Score (OASIS). Data were split into an independent test set (10%) and a ten-fold cross-validation was carried out during training to avoid overfitting. 13,233 distinct variables with heterogeneous data types were included without manual selection or pre-processing. Recordings in the first few hours of a patient's stay were found to be strongly predictive of mortality, outperforming models using SAPS II and OASIS scores within just 2 hours and achieving a state of the art Area Under the Receiver Operating Characteristic (AUROC) value of 0.80 (95% CI 0.79-0.80) at 12 hours vs 0.70 and 0.66 for SAPS II and OASIS at 24 hours respectively. Our model achieves a very strong performance of AUROC 0.86 (95% CI 0.85-0.86) for in-patient mortality prediction after 48 hours on the MIMIC-III dataset. Predictive performance increases over the first 48 hours of the ICU stay, but suffers from diminishing returns, providing rationale for time-limited trials of critical care and suggesting that the timing of decision making can be optimised and individualised.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge