Ari Allyn-Feuer
Fine-tuning Protein Language Models with Deep Mutational Scanning improves Variant Effect Prediction
May 10, 2024
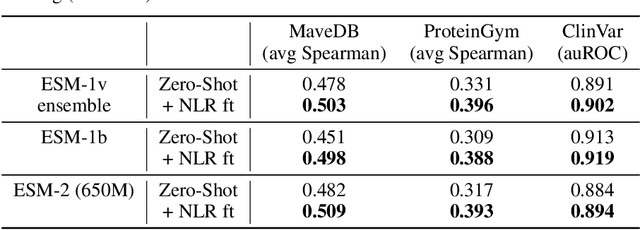

Abstract:Protein Language Models (PLMs) have emerged as performant and scalable tools for predicting the functional impact and clinical significance of protein-coding variants, but they still lag experimental accuracy. Here, we present a novel fine-tuning approach to improve the performance of PLMs with experimental maps of variant effects from Deep Mutational Scanning (DMS) assays using a Normalised Log-odds Ratio (NLR) head. We find consistent improvements in a held-out protein test set, and on independent DMS and clinical variant annotation benchmarks from ProteinGym and ClinVar. These findings demonstrate that DMS is a promising source of sequence diversity and supervised training data for improving the performance of PLMs for variant effect prediction.
Transformative AGI by 2043 is <1% likely
Jun 05, 2023Abstract:This paper is a submission to the Open Philanthropy AI Worldviews Contest. In it, we estimate the likelihood of transformative artificial general intelligence (AGI) by 2043 and find it to be <1%. Specifically, we argue: The bar is high: AGI as defined by the contest - something like AI that can perform nearly all valuable tasks at human cost or less - which we will call transformative AGI is a much higher bar than merely massive progress in AI, or even the unambiguous attainment of expensive superhuman AGI or cheap but uneven AGI. Many steps are needed: The probability of transformative AGI by 2043 can be decomposed as the joint probability of a number of necessary steps, which we group into categories of software, hardware, and sociopolitical factors. No step is guaranteed: For each step, we estimate a probability of success by 2043, conditional on prior steps being achieved. Many steps are quite constrained by the short timeline, and our estimates range from 16% to 95%. Therefore, the odds are low: Multiplying the cascading conditional probabilities together, we estimate that transformative AGI by 2043 is 0.4% likely. Reaching >10% seems to require probabilities that feel unreasonably high, and even 3% seems unlikely. Thoughtfully applying the cascading conditional probability approach to this question yields lower probability values than is often supposed. This framework helps enumerate the many future scenarios where humanity makes partial but incomplete progress toward transformative AGI.
Deep Learning in Pharmacogenomics: From Gene Regulation to Patient Stratification
Mar 07, 2018



Abstract:This Perspective provides examples of current and future applications of deep learning in pharmacogenomics, including: (1) identification of novel regulatory variants located in noncoding domains and their function as applied to pharmacoepigenomics; (2) patient stratification from medical records; and (3) prediction of drugs, targets, and their interactions. Deep learning encapsulates a family of machine learning algorithms that over the last decade has transformed many important subfields of artificial intelligence (AI) and has demonstrated breakthrough performance improvements on a wide range of tasks in biomedicine. We anticipate that in the future deep learning will be widely used to predict personalized drug response and optimize medication selection and dosing, using knowledge extracted from large and complex molecular, epidemiological, clinical, and demographic datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge