Alexandr A. Kalinin
Broad Institute of MIT and Harvard, United States
cubic: CUDA-accelerated 3D Bioimage Computing
Oct 15, 2025Abstract:Quantitative analysis of multidimensional biological images is useful for understanding complex cellular phenotypes and accelerating advances in biomedical research. As modern microscopy generates ever-larger 2D and 3D datasets, existing computational approaches are increasingly limited by their scalability, efficiency, and integration with modern scientific computing workflows. Existing bioimage analysis tools often lack application programmable interfaces (APIs), do not support graphics processing unit (GPU) acceleration, lack broad 3D image processing capabilities, and/or have poor interoperability for compute-heavy workflows. Here, we introduce cubic, an open-source Python library that addresses these challenges by augmenting widely used SciPy and scikit-image APIs with GPU-accelerated alternatives from CuPy and RAPIDS cuCIM. cubic's API is device-agnostic and dispatches operations to GPU when data reside on the device and otherwise executes on CPU, seamlessly accelerating a broad range of image processing routines. This approach enables GPU acceleration of existing bioimage analysis workflows, from preprocessing to segmentation and feature extraction for 2D and 3D data. We evaluate cubic both by benchmarking individual operations and by reproducing existing deconvolution and segmentation pipelines, achieving substantial speedups while maintaining algorithmic fidelity. These advances establish a robust foundation for scalable, reproducible bioimage analysis that integrates with the broader Python scientific computing ecosystem, including other GPU-accelerated methods, enabling both interactive exploration and automated high-throughput analysis workflows. cubic is openly available at https://github$.$com/alxndrkalinin/cubic
cp_measure: API-first feature extraction for image-based profiling workflows
Jul 01, 2025Abstract:Biological image analysis has traditionally focused on measuring specific visual properties of interest for cells or other entities. A complementary paradigm gaining increasing traction is image-based profiling - quantifying many distinct visual features to form comprehensive profiles which may reveal hidden patterns in cellular states, drug responses, and disease mechanisms. While current tools like CellProfiler can generate these feature sets, they pose significant barriers to automated and reproducible analyses, hindering machine learning workflows. Here we introduce cp_measure, a Python library that extracts CellProfiler's core measurement capabilities into a modular, API-first tool designed for programmatic feature extraction. We demonstrate that cp_measure features retain high fidelity with CellProfiler features while enabling seamless integration with the scientific Python ecosystem. Through applications to 3D astrocyte imaging and spatial transcriptomics, we showcase how cp_measure enables reproducible, automated image-based profiling pipelines that scale effectively for machine learning applications in computational biology.
Improving Acne Image Grading with Label Distribution Smoothing
Mar 01, 2024



Abstract:Acne, a prevalent skin condition, necessitates precise severity assessment for effective treatment. Acne severity grading typically involves lesion counting and global assessment. However, manual grading suffers from variability and inefficiency, highlighting the need for automated tools. Recently, label distribution learning (LDL) was proposed as an effective framework for acne image grading, but its effectiveness is hindered by severity scales that assign varying numbers of lesions to different severity grades. Addressing these limitations, we proposed to incorporate severity scale information into lesion counting by combining LDL with label smoothing, and to decouple if from global assessment. A novel weighting scheme in our approach adjusts the degree of label smoothing based on the severity grading scale. This method helped to effectively manage label uncertainty without compromising class distinctiveness. Applied to the benchmark ACNE04 dataset, our model demonstrated improved performance in automated acne grading, showcasing its potential in enhancing acne diagnostics. The source code is publicly available at http://github.com/openface-io/acne-lds.
Ten Quick Tips for Deep Learning in Biology
May 29, 2021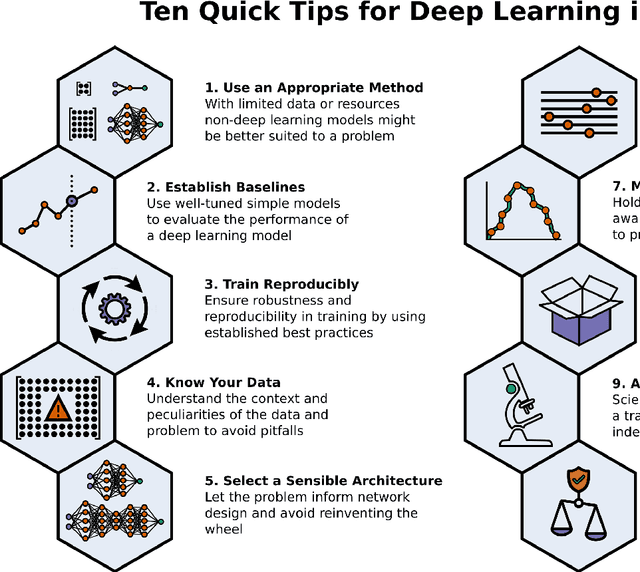
Abstract:Machine learning is a modern approach to problem-solving and task automation. In particular, machine learning is concerned with the development and applications of algorithms that can recognize patterns in data and use them for predictive modeling. Artificial neural networks are a particular class of machine learning algorithms and models that evolved into what is now described as deep learning. Given the computational advances made in the last decade, deep learning can now be applied to massive data sets and in innumerable contexts. Therefore, deep learning has become its own subfield of machine learning. In the context of biological research, it has been increasingly used to derive novel insights from high-dimensional biological data. To make the biological applications of deep learning more accessible to scientists who have some experience with machine learning, we solicited input from a community of researchers with varied biological and deep learning interests. These individuals collaboratively contributed to this manuscript's writing using the GitHub version control platform and the Manubot manuscript generation toolset. The goal was to articulate a practical, accessible, and concise set of guidelines and suggestions to follow when using deep learning. In the course of our discussions, several themes became clear: the importance of understanding and applying machine learning fundamentals as a baseline for utilizing deep learning, the necessity for extensive model comparisons with careful evaluation, and the need for critical thought in interpreting results generated by deep learning, among others.
3D Convolutional Neural Networks for Stalled Brain Capillary Detection
Apr 04, 2021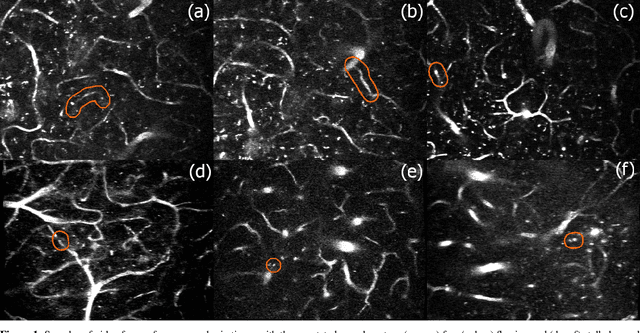


Abstract:Adequate blood supply is critical for normal brain function. Brain vasculature dysfunctions such as stalled blood flow in cerebral capillaries are associated with cognitive decline and pathogenesis in Alzheimer's disease. Recent advances in imaging technology enabled generation of high-quality 3D images that can be used to visualize stalled blood vessels. However, localization of stalled vessels in 3D images is often required as the first step for downstream analysis, which can be tedious, time-consuming and error-prone, when done manually. Here, we describe a deep learning-based approach for automatic detection of stalled capillaries in brain images based on 3D convolutional neural networks. Our networks employed custom 3D data augmentations and were used weight transfer from pre-trained 2D models for initialization. We used an ensemble of several 3D models to produce the winning submission to the Clog Loss: Advance Alzheimer's Research with Stall Catchers machine learning competition that challenged the participants with classifying blood vessels in 3D image stacks as stalled or flowing. In this setting, our approach outperformed other methods and demonstrated state-of-the-art results, achieving 0.85 Matthews correlation coefficient, 85% sensitivity, and 99.3% specificity. The source code for our solution is made publicly available.
Deep Learning for Automatic Pneumonia Detection
May 28, 2020
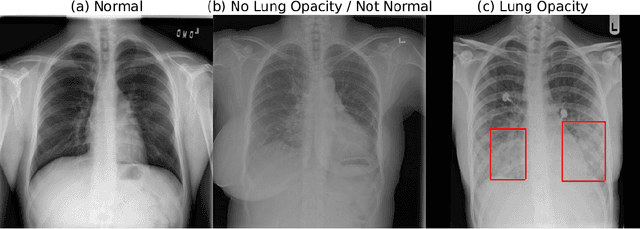
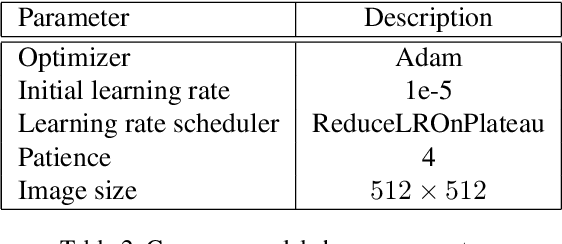
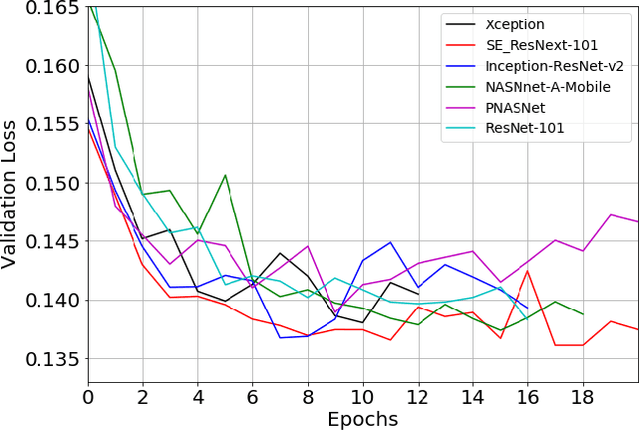
Abstract:Pneumonia is the leading cause of death among young children and one of the top mortality causes worldwide. The pneumonia detection is usually performed through examine of chest X-ray radiograph by highly-trained specialists. This process is tedious and often leads to a disagreement between radiologists. Computer-aided diagnosis systems showed the potential for improving diagnostic accuracy. In this work, we develop the computational approach for pneumonia regions detection based on single-shot detectors, squeeze-and-excitation deep convolution neural networks, augmentations and multi-task learning. The proposed approach was evaluated in the context of the Radiological Society of North America Pneumonia Detection Challenge, achieving one of the best results in the challenge.
Breast Tumor Cellularity Assessment using Deep Neural Networks
May 05, 2019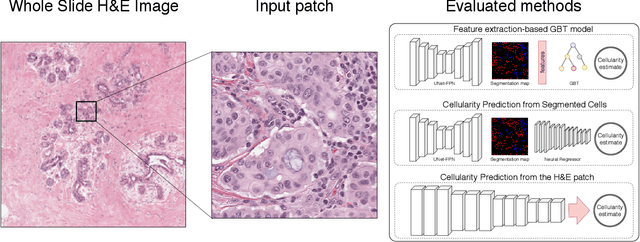
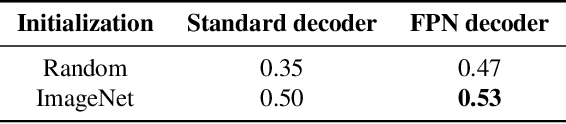
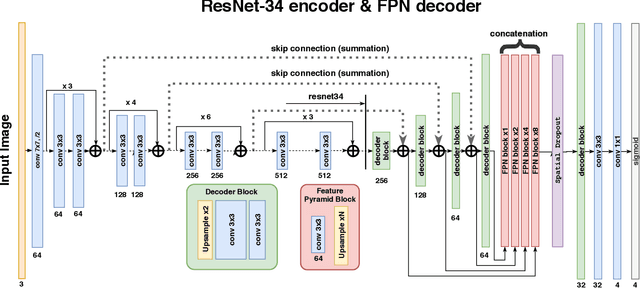
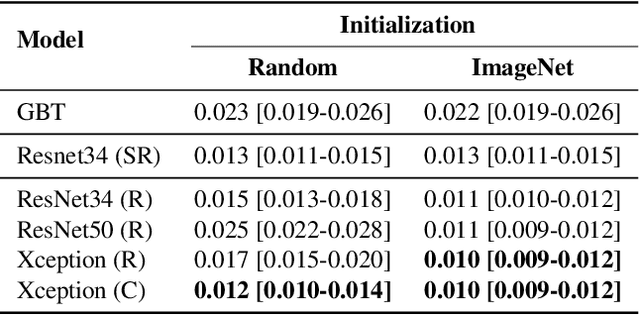
Abstract:Breast cancer is one of the main causes of death worldwide. Histopathological cellularity assessment of residual tumors in post-surgical tissues is used to analyze a tumor's response to a therapy. Correct cellularity assessment increases the chances of getting an appropriate treatment and facilitates the patient's survival. In current clinical practice, tumor cellularity is manually estimated by pathologists; this process is tedious and prone to errors or low agreement rates between assessors. In this work, we evaluated three strong novel Deep Learning-based approaches for automatic assessment of tumor cellularity from post-treated breast surgical specimens stained with hematoxylin and eosin. We validated the proposed methods on the BreastPathQ SPIE challenge dataset that consisted of 2395 image patches selected from whole slide images acquired from 64 patients. Compared to expert pathologist scoring, our best performing method yielded the Cohen's kappa coefficient of 0.70 (vs. 0.42 previously known in literature) and the intra-class correlation coefficient of 0.89 (vs. 0.83). Our results suggest that Deep Learning-based methods have a significant potential to alleviate the burden on pathologists, enhance the diagnostic workflow, and, thereby, facilitate better clinical outcomes in breast cancer treatment.
Albumentations: fast and flexible image augmentations
Sep 18, 2018
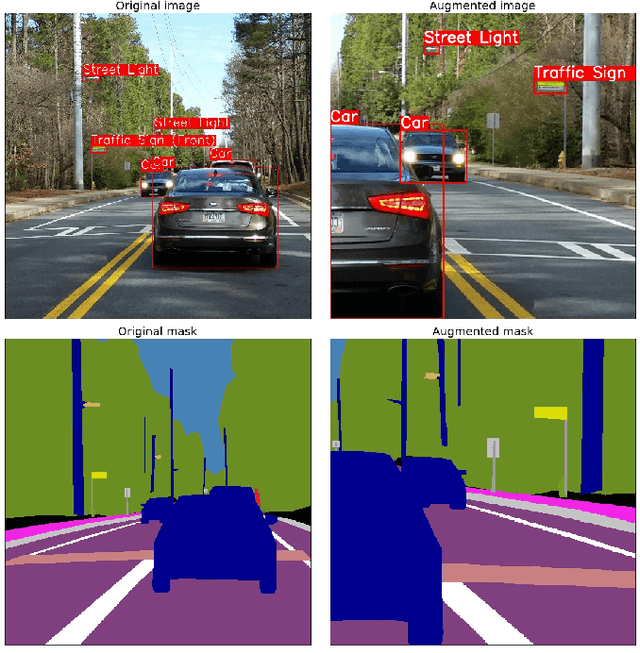

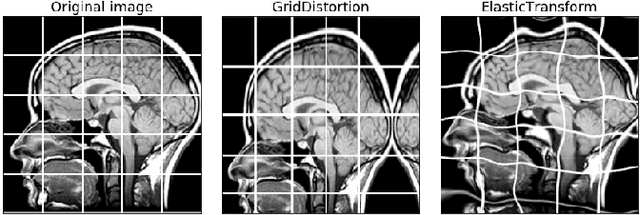
Abstract:Data augmentation is a commonly used technique for increasing both the size and the diversity of labeled training sets by leveraging input transformations that preserve output labels. In computer vision domain, image augmentations have become a common implicit regularization technique to combat overfitting in deep convolutional neural networks and are ubiquitously used to improve performance. While most deep learning frameworks implement basic image transformations, the list is typically limited to some variations and combinations of flipping, rotating, scaling, and cropping. Moreover, the image processing speed varies in existing tools for image augmentation. We present Albumentations, a fast and flexible library for image augmentations with many various image transform operations available, that is also an easy-to-use wrapper around other augmentation libraries. We provide examples of image augmentations for different computer vision tasks and show that Albumentations is faster than other commonly used image augmentation tools on the most of commonly used image transformations. The source code for Albumentations is made publicly available online at https://github.com/albu/albumentations
FPGA Implementation of Convolutional Neural Networks with Fixed-Point Calculations
Aug 29, 2018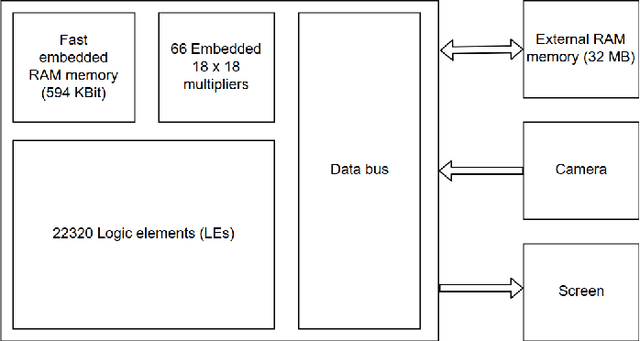
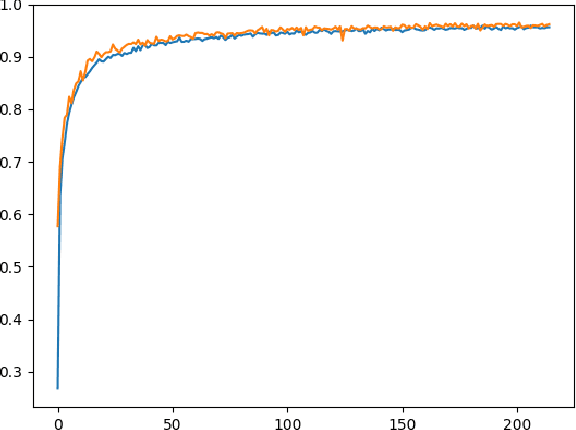
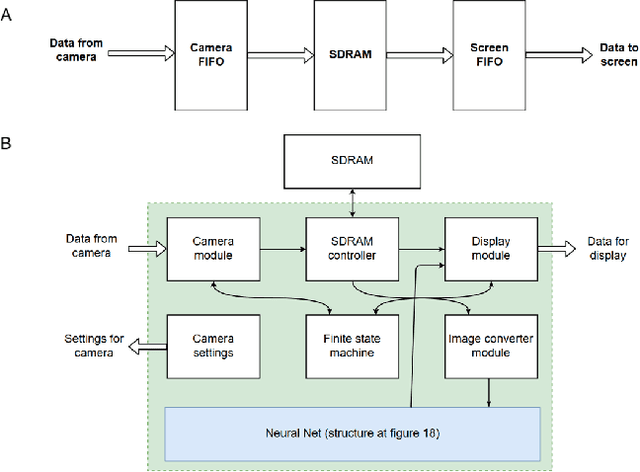
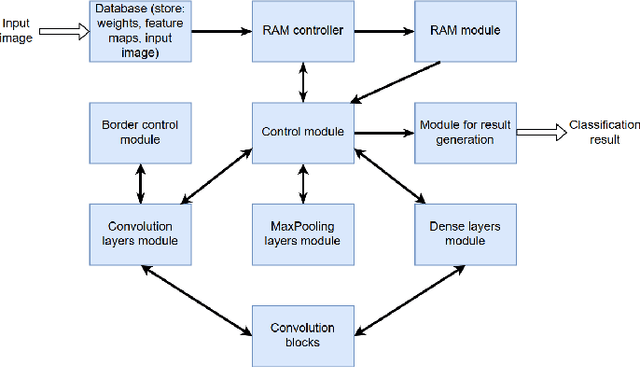
Abstract:Neural network-based methods for image processing are becoming widely used in practical applications. Modern neural networks are computationally expensive and require specialized hardware, such as graphics processing units. Since such hardware is not always available in real life applications, there is a compelling need for the design of neural networks for mobile devices. Mobile neural networks typically have reduced number of parameters and require a relatively small number of arithmetic operations. However, they usually still are executed at the software level and use floating-point calculations. The use of mobile networks without further optimization may not provide sufficient performance when high processing speed is required, for example, in real-time video processing (30 frames per second). In this study, we suggest optimizations to speed up computations in order to efficiently use already trained neural networks on a mobile device. Specifically, we propose an approach for speeding up neural networks by moving computation from software to hardware and by using fixed-point calculations instead of floating-point. We propose a number of methods for neural network architecture design to improve the performance with fixed-point calculations. We also show an example of how existing datasets can be modified and adapted for the recognition task in hand. Finally, we present the design and the implementation of a floating-point gate array-based device to solve the practical problem of real-time handwritten digit classification from mobile camera video feed.
Automatic Instrument Segmentation in Robot-Assisted Surgery Using Deep Learning
Jun 19, 2018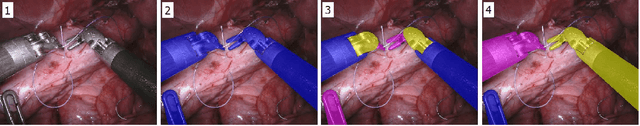
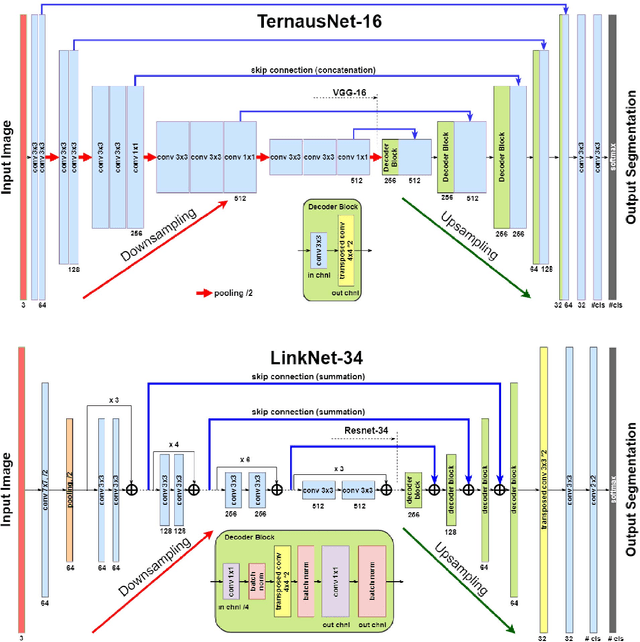
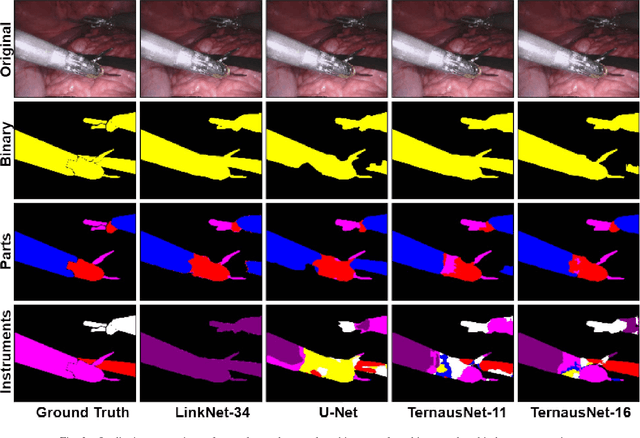
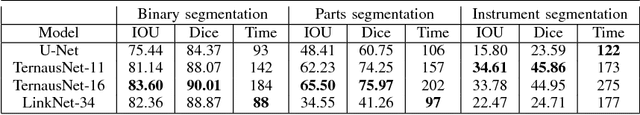
Abstract:Semantic segmentation of robotic instruments is an important problem for the robot-assisted surgery. One of the main challenges is to correctly detect an instrument's position for the tracking and pose estimation in the vicinity of surgical scenes. Accurate pixel-wise instrument segmentation is needed to address this challenge. In this paper we describe our winning solution for MICCAI 2017 Endoscopic Vision SubChallenge: Robotic Instrument Segmentation. Our approach demonstrates an improvement over the state-of-the-art results using several novel deep neural network architectures. It addressed the binary segmentation problem, where every pixel in an image is labeled as an instrument or background from the surgery video feed. In addition, we solve a multi-class segmentation problem, where we distinguish different instruments or different parts of an instrument from the background. In this setting, our approach outperforms other methods in every task subcategory for automatic instrument segmentation thereby providing state-of-the-art solution for this problem. The source code for our solution is made publicly available at https://github.com/ternaus/robot-surgery-segmentation
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge