Marc G. Chevrette
A Graphical Model for Fusing Diverse Microbiome Data
Aug 21, 2022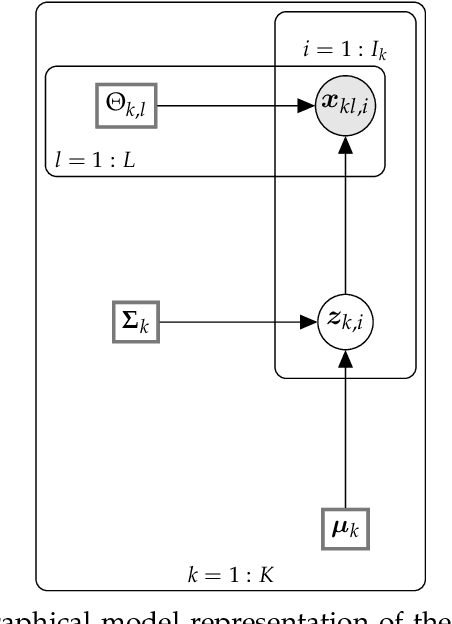
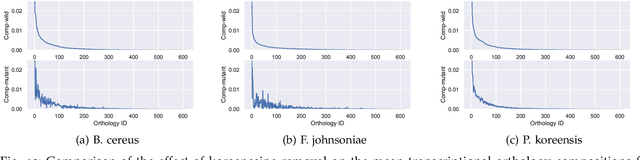
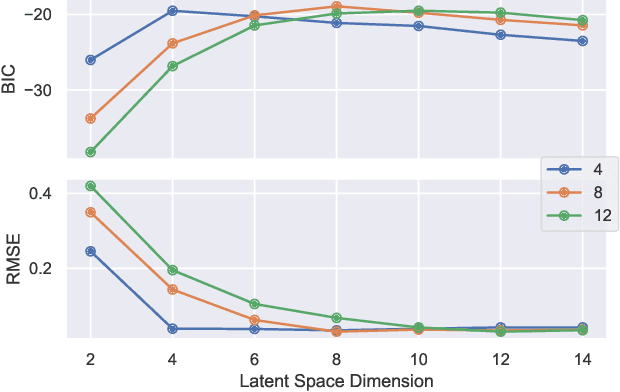

Abstract:This paper develops a Bayesian graphical model for fusing disparate types of count data. The motivating application is the study of bacterial communities from diverse high dimensional features, in this case transcripts, collected from different treatments. In such datasets, there are no explicit correspondences between the communities and each correspond to different factors, making data fusion challenging. We introduce a flexible multinomial-Gaussian generative model for jointly modeling such count data. This latent variable model jointly characterizes the observed data through a common multivariate Gaussian latent space that parameterizes the set of multinomial probabilities of the transcriptome counts. The covariance matrix of the latent variables induces a covariance matrix of co-dependencies between all the transcripts, effectively fusing multiple data sources. We present a computationally scalable variational Expectation-Maximization (EM) algorithm for inferring the latent variables and the parameters of the model. The inferred latent variables provide a common dimensionality reduction for visualizing the data and the inferred parameters provide a predictive posterior distribution. In addition to simulation studies that demonstrate the variational EM procedure, we apply our model to a bacterial microbiome dataset.
Ten Quick Tips for Deep Learning in Biology
May 29, 2021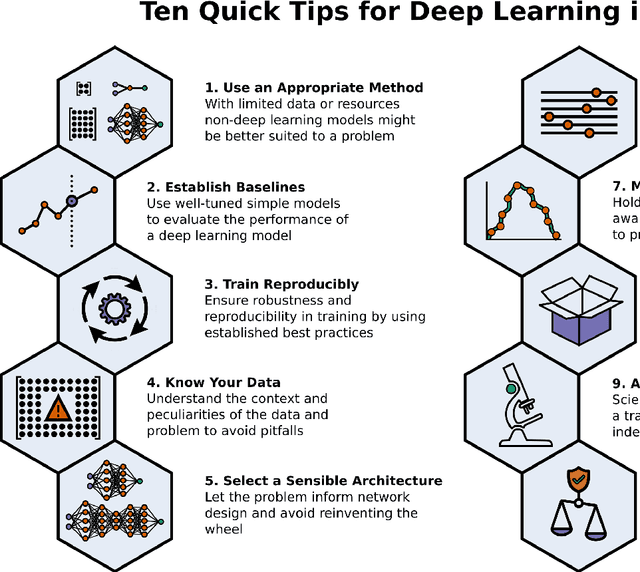
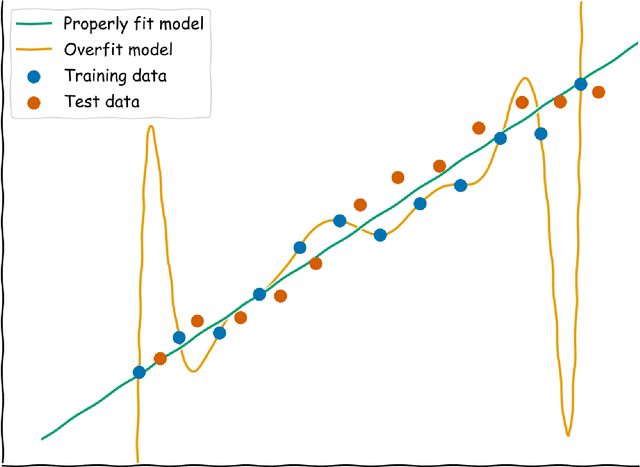
Abstract:Machine learning is a modern approach to problem-solving and task automation. In particular, machine learning is concerned with the development and applications of algorithms that can recognize patterns in data and use them for predictive modeling. Artificial neural networks are a particular class of machine learning algorithms and models that evolved into what is now described as deep learning. Given the computational advances made in the last decade, deep learning can now be applied to massive data sets and in innumerable contexts. Therefore, deep learning has become its own subfield of machine learning. In the context of biological research, it has been increasingly used to derive novel insights from high-dimensional biological data. To make the biological applications of deep learning more accessible to scientists who have some experience with machine learning, we solicited input from a community of researchers with varied biological and deep learning interests. These individuals collaboratively contributed to this manuscript's writing using the GitHub version control platform and the Manubot manuscript generation toolset. The goal was to articulate a practical, accessible, and concise set of guidelines and suggestions to follow when using deep learning. In the course of our discussions, several themes became clear: the importance of understanding and applying machine learning fundamentals as a baseline for utilizing deep learning, the necessity for extensive model comparisons with careful evaluation, and the need for critical thought in interpreting results generated by deep learning, among others.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge