Haonan Zhu
Hierarchical Sparse Bayesian Multitask Model with Scalable Inference for Microbiome Analysis
Feb 04, 2025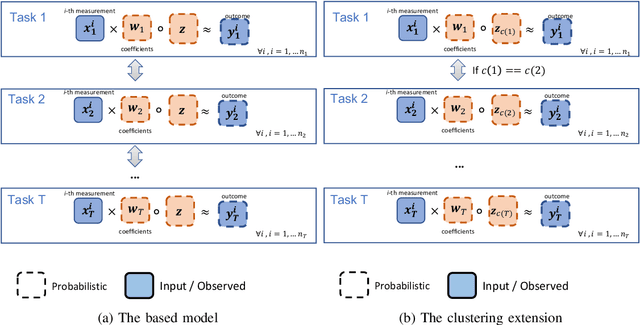
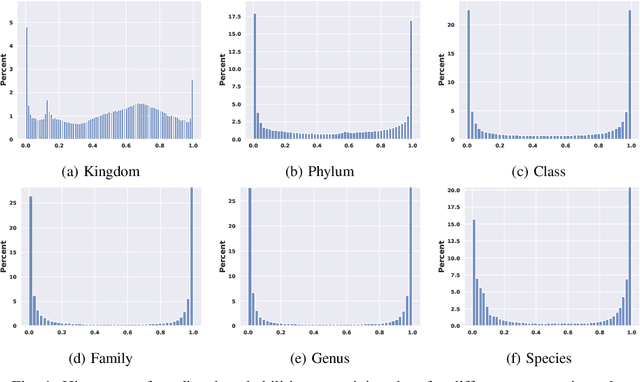
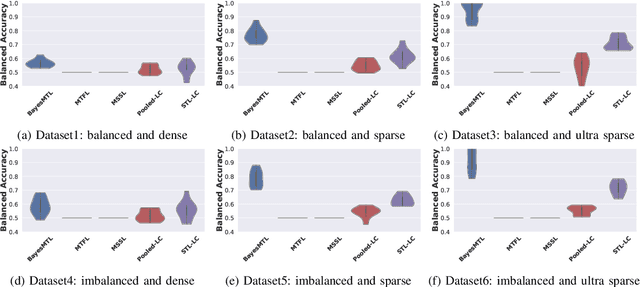
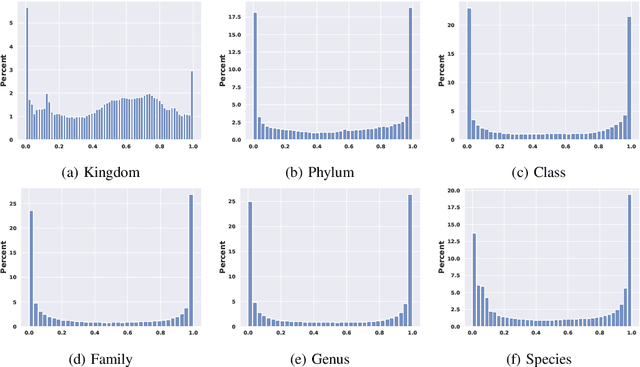
Abstract:This paper proposes a hierarchical Bayesian multitask learning model that is applicable to the general multi-task binary classification learning problem where the model assumes a shared sparsity structure across different tasks. We derive a computationally efficient inference algorithm based on variational inference to approximate the posterior distribution. We demonstrate the potential of the new approach on various synthetic datasets and for predicting human health status based on microbiome profile. Our analysis incorporates data pooled from multiple microbiome studies, along with a comprehensive comparison with other benchmark methods. Results in synthetic datasets show that the proposed approach has superior support recovery property when the underlying regression coefficients share a common sparsity structure across different tasks. Our experiments on microbiome classification demonstrate the utility of the method in extracting informative taxa while providing well-calibrated predictions with uncertainty quantification and achieving competitive performance in terms of prediction metrics. Notably, despite the heterogeneity of the pooled datasets (e.g., different experimental objectives, laboratory setups, sequencing equipment, patient demographics), our method delivers robust results.
Deep Active Learning based Experimental Design to Uncover Synergistic Genetic Interactions for Host Targeted Therapeutics
Feb 03, 2025



Abstract:Recent technological advances have introduced new high-throughput methods for studying host-virus interactions, but testing synergistic interactions between host gene pairs during infection remains relatively slow and labor intensive. Identification of multiple gene knockdowns that effectively inhibit viral replication requires a search over the combinatorial space of all possible target gene pairs and is infeasible via brute-force experiments. Although active learning methods for sequential experimental design have shown promise, existing approaches have generally been restricted to single-gene knockdowns or small-scale double knockdown datasets. In this study, we present an integrated Deep Active Learning (DeepAL) framework that incorporates information from a biological knowledge graph (SPOKE, the Scalable Precision Medicine Open Knowledge Engine) to efficiently search the configuration space of a large dataset of all pairwise knockdowns of 356 human genes in HIV infection. Through graph representation learning, the framework is able to generate task-specific representations of genes while also balancing the exploration-exploitation trade-off to pinpoint highly effective double-knockdown pairs. We additionally present an ensemble method for uncertainty quantification and an interpretation of the gene pairs selected by our algorithm via pathway analysis. To our knowledge, this is the first work to show promising results on double-gene knockdown experimental data of appreciable scale (356 by 356 matrix).
A Graphical Model for Fusing Diverse Microbiome Data
Aug 21, 2022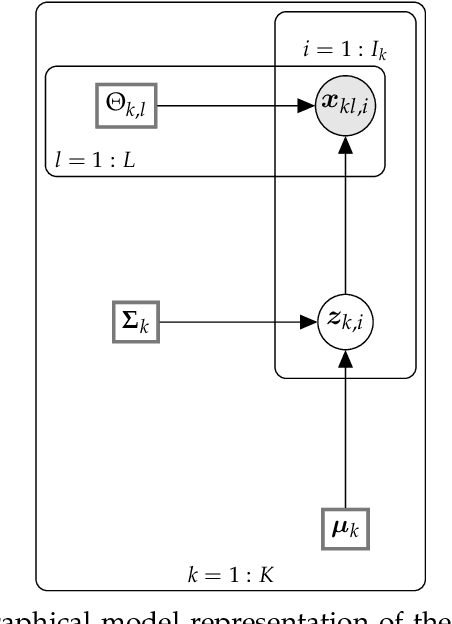
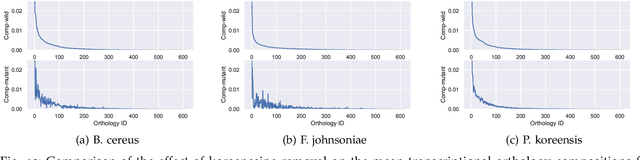
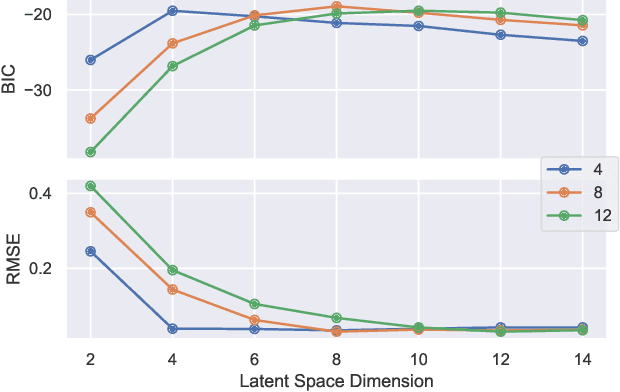

Abstract:This paper develops a Bayesian graphical model for fusing disparate types of count data. The motivating application is the study of bacterial communities from diverse high dimensional features, in this case transcripts, collected from different treatments. In such datasets, there are no explicit correspondences between the communities and each correspond to different factors, making data fusion challenging. We introduce a flexible multinomial-Gaussian generative model for jointly modeling such count data. This latent variable model jointly characterizes the observed data through a common multivariate Gaussian latent space that parameterizes the set of multinomial probabilities of the transcriptome counts. The covariance matrix of the latent variables induces a covariance matrix of co-dependencies between all the transcripts, effectively fusing multiple data sources. We present a computationally scalable variational Expectation-Maximization (EM) algorithm for inferring the latent variables and the parameters of the model. The inferred latent variables provide a common dimensionality reduction for visualizing the data and the inferred parameters provide a predictive posterior distribution. In addition to simulation studies that demonstrate the variational EM procedure, we apply our model to a bacterial microbiome dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge