Braian Peetoom
Deep Active Learning based Experimental Design to Uncover Synergistic Genetic Interactions for Host Targeted Therapeutics
Feb 03, 2025



Abstract:Recent technological advances have introduced new high-throughput methods for studying host-virus interactions, but testing synergistic interactions between host gene pairs during infection remains relatively slow and labor intensive. Identification of multiple gene knockdowns that effectively inhibit viral replication requires a search over the combinatorial space of all possible target gene pairs and is infeasible via brute-force experiments. Although active learning methods for sequential experimental design have shown promise, existing approaches have generally been restricted to single-gene knockdowns or small-scale double knockdown datasets. In this study, we present an integrated Deep Active Learning (DeepAL) framework that incorporates information from a biological knowledge graph (SPOKE, the Scalable Precision Medicine Open Knowledge Engine) to efficiently search the configuration space of a large dataset of all pairwise knockdowns of 356 human genes in HIV infection. Through graph representation learning, the framework is able to generate task-specific representations of genes while also balancing the exploration-exploitation trade-off to pinpoint highly effective double-knockdown pairs. We additionally present an ensemble method for uncertainty quantification and an interpretation of the gene pairs selected by our algorithm via pathway analysis. To our knowledge, this is the first work to show promising results on double-gene knockdown experimental data of appreciable scale (356 by 356 matrix).
Zebra-Llama: A Context-Aware Large Language Model for Democratizing Rare Disease Knowledge
Nov 04, 2024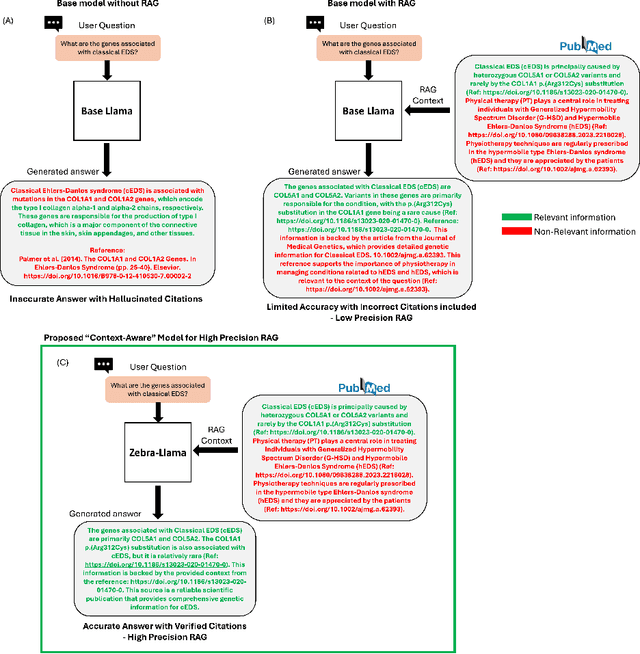
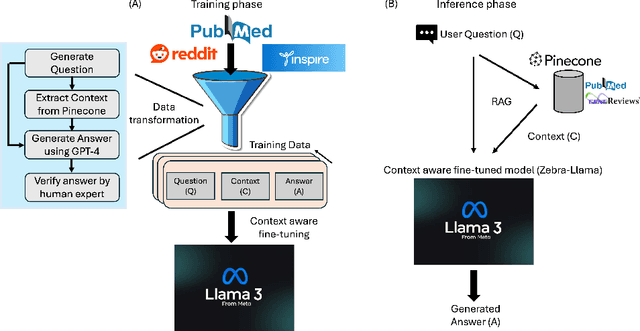
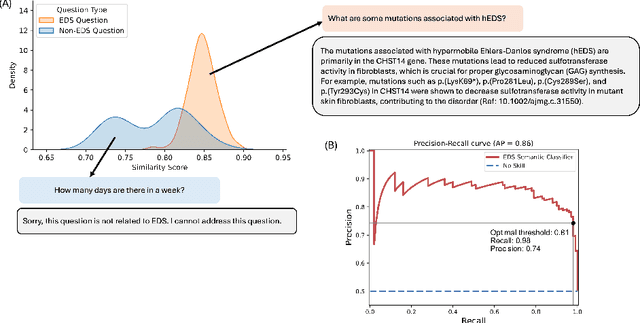
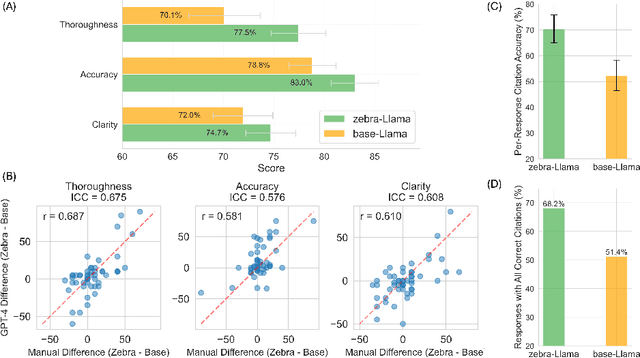
Abstract:Rare diseases present unique challenges in healthcare, often suffering from delayed diagnosis and fragmented information landscapes. The scarcity of reliable knowledge in these conditions poses a distinct challenge for Large Language Models (LLMs) in supporting clinical management and delivering precise patient information underscoring the need for focused training on these 'zebra' cases. We present Zebra-Llama, a specialized context-aware language model with high precision Retrieval Augmented Generation (RAG) capability, focusing on Ehlers-Danlos Syndrome (EDS) as our case study. EDS, affecting 1 in 5,000 individuals, exemplifies the complexities of rare diseases with its diverse symptoms, multiple subtypes, and evolving diagnostic criteria. By implementing a novel context-aware fine-tuning methodology trained on questions derived from medical literature, patient experiences, and clinical resources, along with expertly curated responses, Zebra-Llama demonstrates unprecedented capabilities in handling EDS-related queries. On a test set of real-world questions collected from EDS patients and clinicians, medical experts evaluated the responses generated by both models, revealing Zebra-Llama's substantial improvements over base model (Llama 3.1-8B-Instruct) in thoroughness (77.5% vs. 70.1%), accuracy (83.0% vs. 78.8%), clarity (74.7% vs. 72.0%) and citation reliability (70.6% vs. 52.3%). Released as an open-source resource, Zebra-Llama not only provides more accessible and reliable EDS information but also establishes a framework for developing specialized AI solutions for other rare conditions. This work represents a crucial step towards democratizing expert-level knowledge in rare disease management, potentially transforming how healthcare providers and patients navigate the complex landscape of rare diseases.
Biomedical knowledge graph-enhanced prompt generation for large language models
Nov 29, 2023



Abstract:Large Language Models (LLMs) have been driving progress in AI at an unprecedented rate, yet still face challenges in knowledge-intensive domains like biomedicine. Solutions such as pre-training and domain-specific fine-tuning add substantial computational overhead, and the latter require domain-expertise. External knowledge infusion is task-specific and requires model training. Here, we introduce a task-agnostic Knowledge Graph-based Retrieval Augmented Generation (KG-RAG) framework by leveraging the massive biomedical KG SPOKE with LLMs such as Llama-2-13b, GPT-3.5-Turbo and GPT-4, to generate meaningful biomedical text rooted in established knowledge. KG-RAG consistently enhanced the performance of LLMs across various prompt types, including one-hop and two-hop prompts, drug repurposing queries, biomedical true/false questions, and multiple-choice questions (MCQ). Notably, KG-RAG provides a remarkable 71% boost in the performance of the Llama-2 model on the challenging MCQ dataset, demonstrating the framework's capacity to empower open-source models with fewer parameters for domain-specific questions. Furthermore, KG-RAG enhanced the performance of proprietary GPT models, such as GPT-3.5 which exhibited improvement over GPT-4 in context utilization on MCQ data. Our approach was also able to address drug repurposing questions, returning meaningful repurposing suggestions. In summary, the proposed framework combines explicit and implicit knowledge of KG and LLM, respectively, in an optimized fashion, thus enhancing the adaptability of general-purpose LLMs to tackle domain-specific questions in a unified framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge