Sergio E. Baranzini
Weill Institute for Neurosciences, Department of Neurology, University of California San Francisco, San Francisco, CA, USA
Deep Active Learning based Experimental Design to Uncover Synergistic Genetic Interactions for Host Targeted Therapeutics
Feb 03, 2025



Abstract:Recent technological advances have introduced new high-throughput methods for studying host-virus interactions, but testing synergistic interactions between host gene pairs during infection remains relatively slow and labor intensive. Identification of multiple gene knockdowns that effectively inhibit viral replication requires a search over the combinatorial space of all possible target gene pairs and is infeasible via brute-force experiments. Although active learning methods for sequential experimental design have shown promise, existing approaches have generally been restricted to single-gene knockdowns or small-scale double knockdown datasets. In this study, we present an integrated Deep Active Learning (DeepAL) framework that incorporates information from a biological knowledge graph (SPOKE, the Scalable Precision Medicine Open Knowledge Engine) to efficiently search the configuration space of a large dataset of all pairwise knockdowns of 356 human genes in HIV infection. Through graph representation learning, the framework is able to generate task-specific representations of genes while also balancing the exploration-exploitation trade-off to pinpoint highly effective double-knockdown pairs. We additionally present an ensemble method for uncertainty quantification and an interpretation of the gene pairs selected by our algorithm via pathway analysis. To our knowledge, this is the first work to show promising results on double-gene knockdown experimental data of appreciable scale (356 by 356 matrix).
Beyond Low Earth Orbit: Biomonitoring, Artificial Intelligence, and Precision Space Health
Dec 22, 2021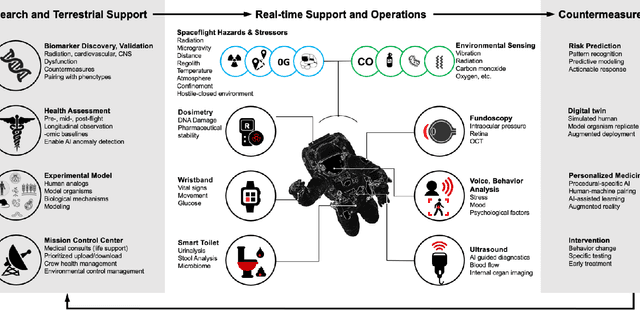
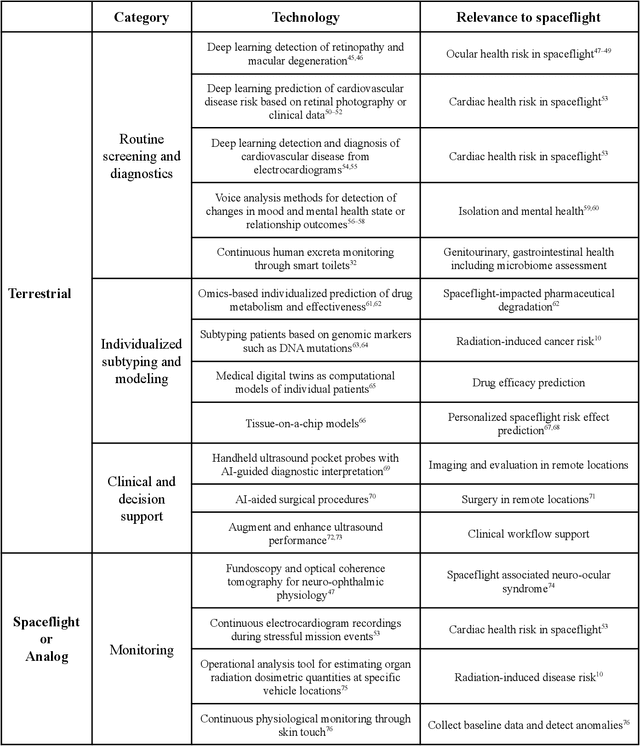
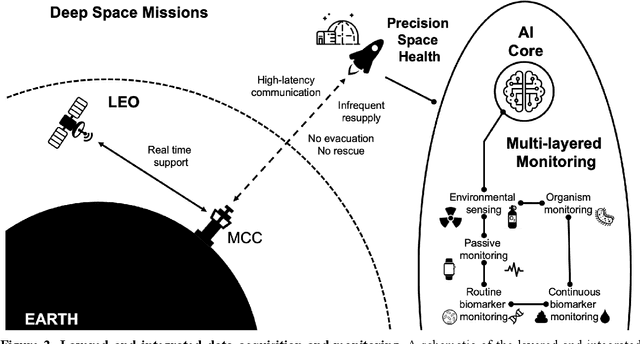
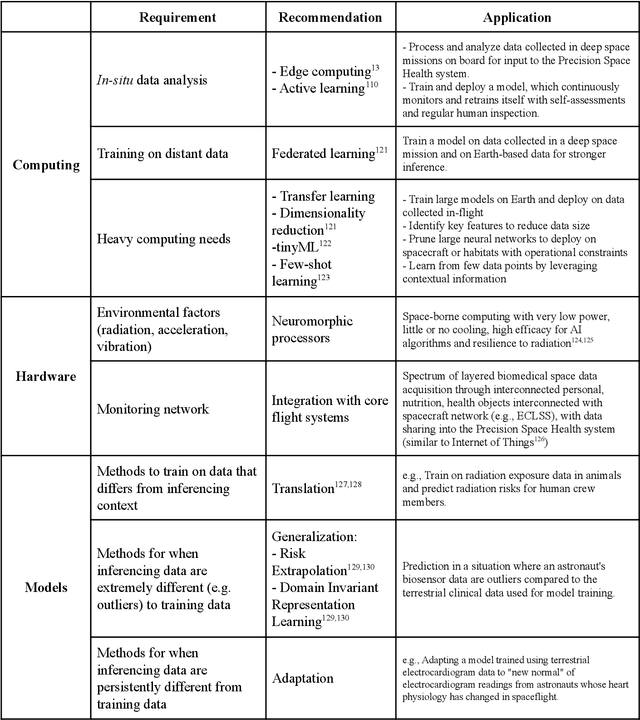
Abstract:Human space exploration beyond low Earth orbit will involve missions of significant distance and duration. To effectively mitigate myriad space health hazards, paradigm shifts in data and space health systems are necessary to enable Earth-independence, rather than Earth-reliance. Promising developments in the fields of artificial intelligence and machine learning for biology and health can address these needs. We propose an appropriately autonomous and intelligent Precision Space Health system that will monitor, aggregate, and assess biomedical statuses; analyze and predict personalized adverse health outcomes; adapt and respond to newly accumulated data; and provide preventive, actionable, and timely insights to individual deep space crew members and iterative decision support to their crew medical officer. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration, on future applications of artificial intelligence in space biology and health. In the next decade, biomonitoring technology, biomarker science, spacecraft hardware, intelligent software, and streamlined data management must mature and be woven together into a Precision Space Health system to enable humanity to thrive in deep space.
Beyond Low Earth Orbit: Biological Research, Artificial Intelligence, and Self-Driving Labs
Dec 22, 2021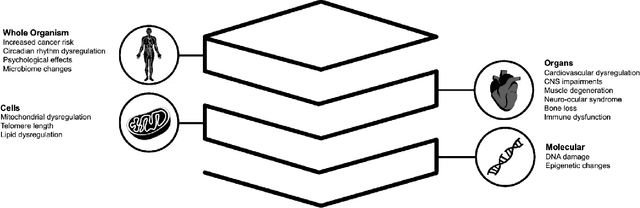
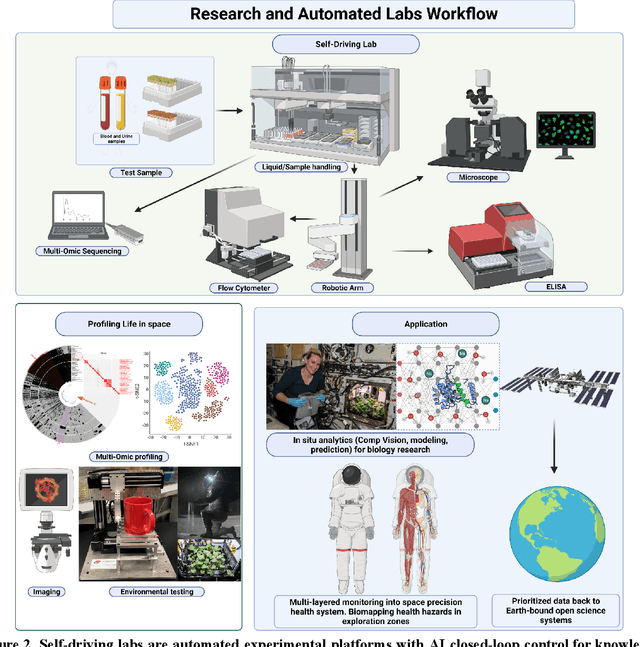
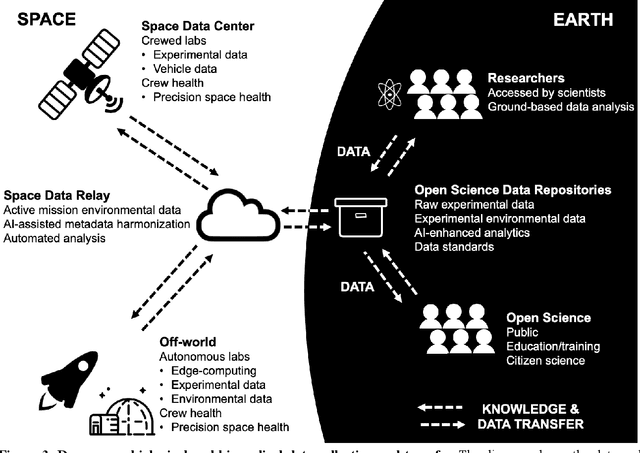
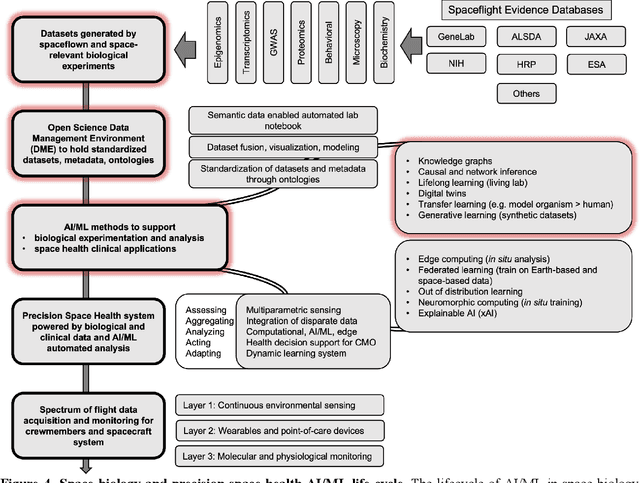
Abstract:Space biology research aims to understand fundamental effects of spaceflight on organisms, develop foundational knowledge to support deep space exploration, and ultimately bioengineer spacecraft and habitats to stabilize the ecosystem of plants, crops, microbes, animals, and humans for sustained multi-planetary life. To advance these aims, the field leverages experiments, platforms, data, and model organisms from both spaceborne and ground-analog studies. As research is extended beyond low Earth orbit, experiments and platforms must be maximally autonomous, light, agile, and intelligent to expedite knowledge discovery. Here we present a summary of recommendations from a workshop organized by the National Aeronautics and Space Administration on artificial intelligence, machine learning, and modeling applications which offer key solutions toward these space biology challenges. In the next decade, the synthesis of artificial intelligence into the field of space biology will deepen the biological understanding of spaceflight effects, facilitate predictive modeling and analytics, support maximally autonomous and reproducible experiments, and efficiently manage spaceborne data and metadata, all with the goal to enable life to thrive in deep space.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge