Jose Cadena
Deep Active Learning based Experimental Design to Uncover Synergistic Genetic Interactions for Host Targeted Therapeutics
Feb 03, 2025



Abstract:Recent technological advances have introduced new high-throughput methods for studying host-virus interactions, but testing synergistic interactions between host gene pairs during infection remains relatively slow and labor intensive. Identification of multiple gene knockdowns that effectively inhibit viral replication requires a search over the combinatorial space of all possible target gene pairs and is infeasible via brute-force experiments. Although active learning methods for sequential experimental design have shown promise, existing approaches have generally been restricted to single-gene knockdowns or small-scale double knockdown datasets. In this study, we present an integrated Deep Active Learning (DeepAL) framework that incorporates information from a biological knowledge graph (SPOKE, the Scalable Precision Medicine Open Knowledge Engine) to efficiently search the configuration space of a large dataset of all pairwise knockdowns of 356 human genes in HIV infection. Through graph representation learning, the framework is able to generate task-specific representations of genes while also balancing the exploration-exploitation trade-off to pinpoint highly effective double-knockdown pairs. We additionally present an ensemble method for uncertainty quantification and an interpretation of the gene pairs selected by our algorithm via pathway analysis. To our knowledge, this is the first work to show promising results on double-gene knockdown experimental data of appreciable scale (356 by 356 matrix).
Scientific Computing Algorithms to Learn Enhanced Scalable Surrogates for Mesh Physics
Apr 01, 2023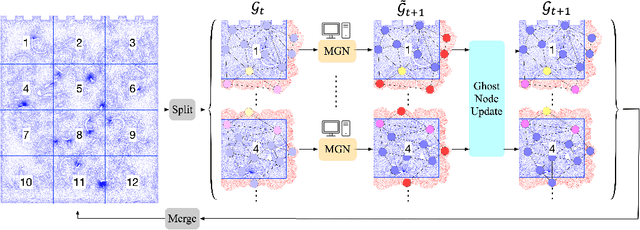
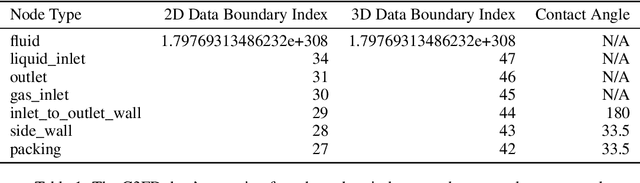
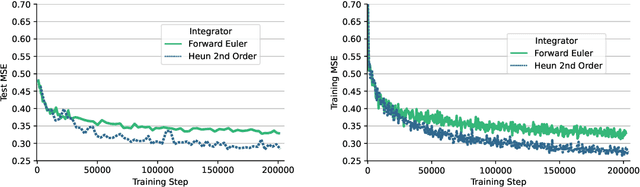
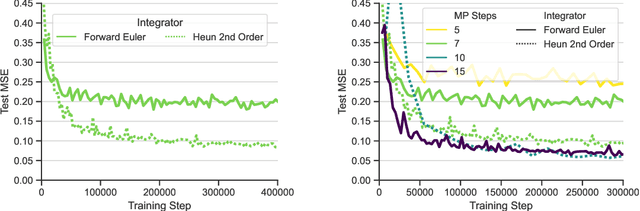
Abstract:Data-driven modeling approaches can produce fast surrogates to study large-scale physics problems. Among them, graph neural networks (GNNs) that operate on mesh-based data are desirable because they possess inductive biases that promote physical faithfulness, but hardware limitations have precluded their application to large computational domains. We show that it is \textit{possible} to train a class of GNN surrogates on 3D meshes. We scale MeshGraphNets (MGN), a subclass of GNNs for mesh-based physics modeling, via our domain decomposition approach to facilitate training that is mathematically equivalent to training on the whole domain under certain conditions. With this, we were able to train MGN on meshes with \textit{millions} of nodes to generate computational fluid dynamics (CFD) simulations. Furthermore, we show how to enhance MGN via higher-order numerical integration, which can reduce MGN's error and training time. We validated our methods on an accompanying dataset of 3D $\text{CO}_2$-capture CFD simulations on a 3.1M-node mesh. This work presents a practical path to scaling MGN for real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge